Data Collection and Preparation - City Digital Twin AR Zoom lens
Skyler Vermeer - Intern Lectorate HTES
Version 1.3.3, March 21st 2023
Version Control
| Version | Date | Author | Amendments | Status |
|---|---|---|---|---|
| 1.0.0 | February 16th 2023 | Skyler Vermeer | Layout + test out downloaded files data | Draft |
| 1.1.0 - 9 | February 17th - March 13th 2023 | Skyler Vermeer | Various variations of code to scrape data from the API | Draft |
| 1.2.0 | March 3rd 2023 | Skyler Vermeer | Exploring 1 location (1 day) | Draft - old version still being updated |
| 1.3.0 | March 14th 2023 | Skyler Vermeer | Combining all data into full datasets | Draft |
| 1.3.1 | March 15th 2023 | Skyler Vermeer | Extend documentation (Sourcing, layout requirement) | Draft |
| 1.3.2 | March 16th 2023 | Skyler Vermeer | Verfication of start dates and units, overall layout and explanation update. | Draft |
| 1.3.3 | March 21st 2023 | Skyler Vermeer | Requirements, Data Quality | Draft |
| 1.3.4 | March 28th 2023 | Skyler Vermeer | Exploring Single day | Draft |
| 1.3.5 | March 30th 2023 | Skyler Vermeer | Exploring Single day - continued | Draft |
The first thing we do with any exploratory data analysis, is get our basic packages imported and print their versions so it is transparent what we did and can be reproduced.
import os
from datetime import datetime
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
print('pandas version:', pd.__version__)
print('numpy version:', np.__version__)
print('matplotlib version:', matplotlib.__version__)
pandas version: 1.5.2
numpy version: 1.23.0
matplotlib version: 3.6.2
Data Sourcing
To obtain data that is relevant to my Fine Dust domain and can help with anwering my subquestions: “What data is available regarding this problem?” and “How can AI be used to make predictions using this data?”, I went to the Open Eindhoven Data Eindhoven Open Data. I did this, because the City Digital Twin project works on the basis of creating a Digital Twin of the City Eindhoven using the open data that the city shares. Here I searched for all the datasets relevant to fine dust.
Different datasets based on fine dust on platform.
- Real-time fine dust monitoring {SOURCE}, it contains real-time data of fine dust in Eindhoven, updated every hour and collected every 10 minutes. It is collected over just under 50 locations in and around the city Eindhoven. It is made to give an overview of the air quality in South-East Brabant. It measures PM1, PM2.5, PM10 and NO2. It has locations of varying concentrations of fine dust particles. They give you the ability to download the data, select specific locations and download in batches of up to 70 days. It also has an API {SOURCE} where you can obtain historical data, or search for specific moments, and filter what features you want.
- Locaties Caireboxen Fijnstof-monitoring {SOURCE}, it contains the locations of the Fine Dust monitoring boxes, in the Eindhoven municipality. It has the ID, name, town and street, observables, API URL and location of the monitoring boxes.
During my research I was referred to some datasets for additional information or data
- SamenMeten {SOURCE}, contains data collected by a variety of organizations and is combined by RIVM into one data portal. Most sensors measure multiple times an hour, and air quality is averaged each hour. The measurements are calibrated with the use of data science, to correct unwanted influences such as humidity. Raw sensormeasurements are also available.
- WorldClim {SOURCE}, contains climate data for 1970-2000. There is monthly climate data for various weather features, as well as 19 bioclimatic variables. It is available at 4 spatial resulations, between 30 seconds to 10 minutes. There is also monthly weather data from 1960-2018 regarding the minimum and maximum temperature and precipation. Based on this, a model was created for future weather and climate predictions.
Based on the research, and the data we have available, we are going to visualize the current fine dust data and predict the amount of fine dust (PM1, PM2.5 and PM10) in the future based on historical data and if possible also incorporating weather and climate. This, because it fits the requests of the client, helps with answering the research question and the data allows for it. The weather data is added as based on my domain research, the weather is an important aspect in the behaviour of fine dust, so if possible we want to incorporate this. There are other prediction opportunities, such as classifying the type of fine dust, whether it is harmful or not, however that is not possible within the current data.
Data requirements
For this project we want to create a prediction of the amount of fine dust in the air at a certain moment in the future. We also want to show the current amount of fine dust in the area. To do this, we will need a few things from our data.
- The data must be collected consistently, so we can see trends and outliers.
- The data must be collected regularly, so we can show the current amount of fine dust in the area.
- The data must contain some form of timestamp.
- The data must contain some form of location.
- The data must contain a measurement of fine dust, preferably PM1, PM2.5 and PM10.
Based on this, we will be using the real-time fine dust monitoring that was shared on the Eindhoven Open Data platform. We will use the Locaties Caireboxen fijnstof monitoring to verify the locations that are being monitored.
If we want, we could use WorldClim to incorporate weather and climate data into our prediction. However, for now we will first explore the fine dust monitoring dataset and work from there.
Collecting the Data
Downloading
To start this project, I manually downloaded data from December 21st to February 16th 2023 due to the 70 day maximum. I stored the manually downloaded files in a directory called csv_files/manual_download for ease of access.
manual_file_path = "C:/Users/Public/Documents/Github/Internship-S5/cdt-a-i-r-zoom-lens-back-end/csv_files/manual_download/"
Let’s take a look at what this data looks like
pd.read_csv(manual_file_path + 'Dustmonitoring data.2022-12-21.2023-02-16.csv', sep=';')
| Unnamed: 0 | Unnamed: 1 | I01 | I01.1 | I01.2 | I01.3 | I01.4 | I01.5 | I02 | I02.1 | ... | I57.2 | I57.3 | I57.4 | I57.5 | I58 | I58.1 | I58.2 | I58.3 | I58.4 | I58.5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Time | Time | Lat | Lon | PM1 | PM2.5 | PM10 | NO2 | Lat | Lon | ... | PM1 | PM2.5 | PM10 | NO2 | Lat | Lon | PM1 | PM2.5 | PM10 | NO2 |
| 1 | UTC | Local | ° | ° | μg/m³ | μg/m³ | μg/m³ | μg/m³\r\n | ° | ° | ... | μg/m³ | μg/m³ | μg/m³ | μg/m³\r\n | ° | ° | μg/m³ | μg/m³ | μg/m³ | μg/m³\r\n |
| 2 | 2022-12-21 00:00:00 | 2022-12-21 01:00:00 | 51,4063 | 5,4574 | 17,21 | 19,13 | 30,32 | 22 | NaN | NaN | ... | 10,44 | 10,71 | 20,22 | 11 | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | 2022-12-21 00:00:01 | 2022-12-21 01:00:01 | NaN | NaN | NaN | NaN | NaN | NaN | 51,4379 | 5,3581 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | 2022-12-21 00:00:04 | 2022-12-21 01:00:04 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | 51,4904 | 5,3941 | 15,75 | 16,54 | 17,94 | 15 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 28915 | 2023-02-16 23:40:04 | 2023-02-17 00:40:04 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | 51,4904 | 5,3941 | 11,01 | 11,85 | 13,49 | 13.0 |
| 28916 | 2023-02-16 23:49:59 | 2023-02-17 00:49:59 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 28917 | 2023-02-16 23:50:00 | 2023-02-17 00:50:00 | 51,4063 | 5,4573 | 15,00 | 16,16 | 17,20 | 2.0 | 51,4379 | 5,3582 | ... | 9,80 | 10,20 | 24,09 | 16.0 | NaN | NaN | NaN | NaN | NaN | NaN |
| 28918 | 2023-02-16 23:50:01 | 2023-02-17 00:50:01 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 28919 | 2023-02-16 23:50:04 | 2023-02-17 00:50:04 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | 51,4904 | 5,3941 | 10,48 | 11,02 | 11,56 | 13.0 |
28920 rows × 287 columns
We can see that the information is quite wonky. It is technically the information, but it has a column for every location and measurement. The columns that are not related to the location of the entry, are NaN. It would need a lot of adjusting to gather the info, not really a good idea. Let’s look at a selection of a single location
pd.read_csv(manual_file_path + 'Dustmonitoring data.2022-12-21.2023-02-16_I01.csv', sep=';')
| Unnamed: 0 | Unnamed: 1 | I01 | I01.1 | I01.2 | I01.3 | I01.4 | I01.5 | |
|---|---|---|---|---|---|---|---|---|
| 0 | Time | Time | Lat | Lon | PM1 | PM2.5 | PM10 | NO2 |
| 1 | UTC | Local | ° | ° | μg/m³ | μg/m³ | μg/m³ | μg/m³\r\n |
| 2 | 2022-12-21 00:00:00 | 2022-12-21 01:00:00 | 51,4063 | 5,4574 | 17,21 | 19,13 | 30,32 | 22 |
| 3 | 2022-12-21 00:10:00 | 2022-12-21 01:10:00 | 51,4063 | 5,4574 | 18,29 | 20,75 | 25,83 | 25 |
| 4 | 2022-12-21 00:20:01 | 2022-12-21 01:20:01 | 51,4063 | 5,4574 | 18,88 | 22,04 | 32,27 | 25 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 8340 | 2023-02-16 23:10:00 | 2023-02-17 00:10:00 | 51,4063 | 5,4573 | 19,13 | 21,14 | 29,29 | 2 |
| 8341 | 2023-02-16 23:20:00 | 2023-02-17 00:20:00 | 51,4063 | 5,4573 | 18,37 | 20,23 | 32,35 | 2 |
| 8342 | 2023-02-16 23:30:00 | 2023-02-17 00:30:00 | 51,4063 | 5,4573 | 17,36 | 19,13 | 26,48 | 2 |
| 8343 | 2023-02-16 23:40:01 | 2023-02-17 00:40:01 | 51,4063 | 5,4573 | 18,19 | 19,54 | 25,84 | 2 |
| 8344 | 2023-02-16 23:50:00 | 2023-02-17 00:50:00 | 51,4063 | 5,4573 | 15,00 | 16,16 | 17,20 | 2 |
8345 rows × 8 columns
This would work as it just needs adjustment of the top 2 rows into the column name. However, manually downloading it would take too much work because 58 locations in batches of 70 days would be around 580+ requests. There is also an API. The api works with odata, which if you use the correct query will also provide you with @odata.nextlink which will give you the query to get even more of the same query aside from the data you have already gotten. This means that if we can manipulate this query to automatically process all the information, we can scrape the data in a loop (and also in the future starting from a specific point)
https://api.dustmonitoring.nl/v1/project(30)/location(521)/stream I found that this gives the data for location ‘521’, and gives an @odata.nextlink.
Using the API
So first I am going to get all the available locations.
import requests as request
res = request.get("https://api.dustmonitoring.nl/v1/project(30)/locations")
data = res.json()
locations = []
for item in data['value']:
locations.append(item['ID'])
Then it is time to obtain the info from all these locations.
print(locations)
print(len(locations))
[531, 521, 542, 543, 553, 544, 545, 532, 533, 554, 534, 535, 546, 536, 556, 522, 557, 547, 549, 524, 537, 525, 526, 539, 551, 540, 558, 527, 528, 529, 530, 560, 561, 562, 563, 564, 565, 566, 567, 568, 569, 570, 571, 572, 573, 574, 575, 576, 577]
49
file_path = "C:/Users/Public/Documents/Github/Internship-S5/cdt-a-i-r-zoom-lens-back-end/csv_files/raw_data/"
file_list = os.listdir(file_path)
for location in locations:
request_string = "https://api.dustmonitoring.nl/v1/project(30)/location(" + str(location) + ")/stream"
index = 0
dataframes = []
while index != -1:
res = request.get(request_string)
data = res.json()
df = pd.DataFrame(data['value'])
dataframes.append(df)
try:
request_string = data['@odata.nextLink']
except KeyError:
index = -1
print(request_string)
combined_df = pd.DataFrame()
for dataframe in dataframes:
combined_df = combined_df.append(dataframe)
combined_df.to_csv(file_path + str(location) + '_raw_data')
print("done")
# This code block is to save a location if the last step (saving) threw an error for whatever reason
# combined_df.to_csv(file_path + str(577) + '_raw_data')
# This code block is to finish a location if it was interrupted while scraping so it doesn't have to go from the start.
# while index != -1:
# res = request.get(request_string)
# data = res.json()
# df = pd.DataFrame(data['value'])
# dataframes.append(df)
# try:
# request_string = data['@odata.nextLink']
# except KeyError:
# index = -1
# print(request_string)
# combined_df = pd.DataFrame()
# for dataframe in dataframes:
# combined_df = combined_df.append(dataframe)
#
# combined_df.to_csv(file_path + str(location) + '_raw_data')
The next piece of code is to update the dataframe since the initial scraping, so it can be combined with KNMI data. It is practically the same, but it updates from the last known data point.
current_data = pd.read_csv(file_path + str(531) + "_raw_data")
current_data.tail(1).Timestamp.values[0]
'2023-05-19T09:00:00Z'
for location in locations:
current_data = pd.read_csv(file_path + str(location) + "_raw_data")
last_datetime = current_data.tail(1).Timestamp.values[0]
request_string = "https://api.dustmonitoring.nl/v1/project(30)/location(" + str(location) + ")/stream?$filter=timestamp gt " + last_datetime
index = 0
dataframes = []
while index != -1:
res = request.get(request_string)
data = res.json()
df = pd.DataFrame(data['value'])
dataframes.append(df)
try:
request_string = data['@odata.nextLink']
except KeyError:
index = -1
print(request_string)
combined_df = current_data
for dataframe in dataframes:
combined_df = pd.concat([combined_df, dataframe], ignore_index=True)
combined_df.to_csv(file_path + str(location) + '_raw_data')
print("done")
Preparation
Single location
Now that we have all the data, it is time to combine the raw data into a large dataset. But first, let’s look at a single location dataset.
df_521 = pd.read_csv(file_path + '521_raw_data')
print(df_521.sample(5))
Unnamed: 0.4 Unnamed: 0.3 Unnamed: 0.2 Unnamed: 0.1 Unnamed: 0 \
86328 86328 86328.0 86328.0 86328.0 72.0
16279 16279 16279.0 16279.0 16279.0 7.0
262576 262576 262576.0 262576.0 262576.0 64.0
607332 607332 607332.0 607332.0 607332.0 NaN
585241 585241 585241.0 585241.0 585241.0 25.0
Timestamp Location Entity Name Value \
86328 2021-06-13T16:00:00Z 521 129 PM2.5 8.3600
16279 2021-02-09T09:30:00Z 521 130 PM10 12.1900
262576 2022-01-30T05:20:00Z 521 10 Lat 51.3468
607332 2023-03-12T07:20:01Z 521 145 UFP 11782.0000
585241 2023-02-16T16:40:01Z 521 10 Lat 51.4379
Unit
86328 μg/m³
16279 μg/m³
262576 °
607332 counts/cm³\r\n
585241 °
We see we start with an unnamed column, which is because the dataframes were combined including their index, so we need to remove that. It would also be useful if we seperate the timestamp into date and time. It might be useful to combine the name and unit as that is just useful for reference. Or create a separate dataframe to reference it. It is also useful to seperate the Lon/Lat values from the rest of the values as even though the values are there, and we need them, they are somewhat the same per location with a few inaccuracies in measuring. So we should save the locations in a separate csv, and make it one per location. Entity is the ID associated with a measurement, so all Lat measurements have entity 10 and all PM10 have entity 130. Also in units some of them have \r\n attached to them, which isn’t part of the unit.
First, let’s remove the extra, unnamed index column.
print(df_521.loc[:, 'Timestamp':'Unit'].sample(5))
Timestamp Location Entity Name Value Unit
48853 2021-04-22T08:00:00Z 521 10 Lat 51.3468 °
131742 2021-08-16T00:00:00Z 521 129 PM2.5 9.8400 μg/m³
313889 2022-03-30T21:30:00Z 521 10 Lat 51.3468 °
661700 2023-05-10T19:10:00Z 521 128 PM1 9.2100 μg/m³
388315 2022-07-22T23:10:01Z 521 11 Lon 4.7966 °
Now, let’s separate the values. The names are short enough that they can easily be used as an index. So we want to split the data into three datasets. A dataset with the different locations (ID combined with Lat and Lon), a dataset with the different measurements (Entity ID, Name, and Unit), and a dataset with the main data we will be using, which will be a separate column for day and time, a location id, a name , and a measurement.
Let’s check first whether there is one unique unit per measurement or whether there are multiple.
df_521.groupby('Name')['Unit'].unique()
Name
Lat [°]
Lon [°]
NO2 [μg/m³\r\n]
PM1 [μg/m³]
PM10 [μg/m³]
PM2.5 [μg/m³]
UFP [counts/cm³\r\n]
Name: Unit, dtype: object
It doesn’t look like it. Of course this is just one location, and we need to check with the others, but it is positive that at least for this location it is just a static combination. Let’s just save all this as a static dataframe for now, and we can verify whether it is the same with all locations later.
units = df_521.groupby('Name')['Unit'].unique()
entities = df_521.groupby('Name')['Entity'].unique()
measurement_info = pd.DataFrame()
units[2][0] = units[2][0][:-2]
units[6][0] = units[6][0][:-2]
for idx in range(len(units)):
measurement_info = pd.concat(
[measurement_info, pd.DataFrame([units.index[idx], units[idx][0], entities[idx][0]]).T])
measurement_info.columns = ['Name', 'Unit', 'Entity_ID']
measurement_info = measurement_info.reset_index(drop=True)
print(measurement_info)
Name Unit Entity_ID
0 Lat ° 10
1 Lon ° 11
2 NO2 μg/m³ 146
3 PM1 μg/m³ 128
4 PM10 μg/m³ 130
5 PM2.5 μg/m³ 129
6 UFP counts/cm³ 145
measurement_info.to_csv(
'C:/Users/Public/Documents/Github/Internship-S5/cdt-a-i-r-zoom-lens-back-end/csv_files/' + 'measurement_info',
index=False)
Now, let’s see about the locations. We know that though the there is only one correct longitude and latitude combination per location, due to some errors there are sometimes other locations. However, there is also the dataset that is saved in ‘locaties-airboxen.csv’ which contains the actual locations of the sensor.
airboxes = pd.read_csv('csv_files/locaties-airboxen.csv', sep=';')
airbox_521 = airboxes.loc[airboxes.ID == 521]
airbox_521
| ID | Name | Lat | Lon | Description | Observables/0 | Observables/1 | Observables/2 | Observables/3 | Observables/4 | URL | geopoint | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 521 | I02 | 51.437895 | 5.358142 | Eindhoven, Scherpenering | Lat | Lon | PM1 | PM2.5 | PM10 | https://api.dustmonitoring.nl/v1/project(30)/l... | 51.437895, 5.358142 |
lon = airbox_521['Lon']
lat = airbox_521['Lat']
print(str(521) + ', Lat: ' + str(lat.values[0]) + ', Lon: ' + str(lon.values[0]))
521, Lat: 51.437895, Lon: 5.358142
Now we know how to easily get more data about the different locations. The last and most important aspect that we need to get is the data that we want to use. For that we want a separate column for day and time, a location id, a name , and a measurement.
measurement_521_data = df_521.loc[:, ['Timestamp', 'Location', 'Name', 'Value']]
measurement_521_drop_unused = measurement_521_data[
(measurement_521_data.Name != 'Lat') & (measurement_521_data.Name != 'Lon') & (
measurement_521_data.Name != 'NO2') & (measurement_521_data.Name != 'UFP')].copy()
print(measurement_521_drop_unused)
Timestamp Location Name Value
54 2021-01-21T13:00:00Z 521 PM10 22.03
55 2021-01-21T13:00:00Z 521 PM2.5 10.54
56 2021-01-21T13:00:00Z 521 PM1 7.29
59 2021-01-21T13:10:00Z 521 PM10 20.90
60 2021-01-21T13:10:00Z 521 PM2.5 10.52
... ... ... ... ...
670192 2023-05-19T08:50:00Z 521 PM2.5 13.44
670193 2023-05-19T08:50:00Z 521 PM10 19.53
670198 2023-05-19T09:00:00Z 521 PM1 8.15
670199 2023-05-19T09:00:00Z 521 PM2.5 11.33
670200 2023-05-19T09:00:00Z 521 PM10 26.28
[335613 rows x 4 columns]
measurement_521_drop_unused['Timestamp'] = pd.to_datetime(measurement_521_drop_unused['Timestamp'])
measurement_521_drop_unused['Date'] = [d.date() for d in measurement_521_drop_unused['Timestamp']]
measurement_521_drop_unused['Time'] = [d.time() for d in measurement_521_drop_unused['Timestamp']]
measurement_521_drop_unused.head(5)
| Timestamp | Location | Name | Value | Date | Time | |
|---|---|---|---|---|---|---|
| 54 | 2021-01-21 13:00:00+00:00 | 521 | PM10 | 22.03 | 2021-01-21 | 13:00:00 |
| 55 | 2021-01-21 13:00:00+00:00 | 521 | PM2.5 | 10.54 | 2021-01-21 | 13:00:00 |
| 56 | 2021-01-21 13:00:00+00:00 | 521 | PM1 | 7.29 | 2021-01-21 | 13:00:00 |
| 59 | 2021-01-21 13:10:00+00:00 | 521 | PM10 | 20.90 | 2021-01-21 | 13:10:00 |
| 60 | 2021-01-21 13:10:00+00:00 | 521 | PM2.5 | 10.52 | 2021-01-21 | 13:10:00 |
Now we have successfully split the Date and Time column, it is time to start converting the data to have all the different values in one single column.
df_521_measurements = pd.DataFrame(columns=['Date', 'Time', 'Location', 'PM10', 'PM2.5', 'PM1'])
date_time_groups = measurement_521_drop_unused.groupby(['Date', 'Time'])
groupings = [date_time_groups.get_group(x) for x in date_time_groups.groups]
print(groupings[:5])
print(groupings[-5:])
[ Timestamp Location Name Value Date Time
54 2021-01-21 13:00:00+00:00 521 PM10 22.03 2021-01-21 13:00:00
55 2021-01-21 13:00:00+00:00 521 PM2.5 10.54 2021-01-21 13:00:00
56 2021-01-21 13:00:00+00:00 521 PM1 7.29 2021-01-21 13:00:00, Timestamp Location Name Value Date Time
59 2021-01-21 13:10:00+00:00 521 PM10 20.90 2021-01-21 13:10:00
60 2021-01-21 13:10:00+00:00 521 PM2.5 10.52 2021-01-21 13:10:00
61 2021-01-21 13:10:00+00:00 521 PM1 7.02 2021-01-21 13:10:00, Timestamp Location Name Value Date Time
64 2021-01-21 13:20:00+00:00 521 PM10 21.30 2021-01-21 13:20:00
65 2021-01-21 13:20:00+00:00 521 PM2.5 10.93 2021-01-21 13:20:00
66 2021-01-21 13:20:00+00:00 521 PM1 6.66 2021-01-21 13:20:00, Timestamp Location Name Value Date Time
69 2021-01-21 13:30:00+00:00 521 PM10 20.23 2021-01-21 13:30:00
70 2021-01-21 13:30:00+00:00 521 PM2.5 11.72 2021-01-21 13:30:00
71 2021-01-21 13:30:00+00:00 521 PM1 7.67 2021-01-21 13:30:00, Timestamp Location Name Value Date Time
74 2021-01-21 13:40:00+00:00 521 PM10 19.25 2021-01-21 13:40:00
75 2021-01-21 13:40:00+00:00 521 PM2.5 11.37 2021-01-21 13:40:00
76 2021-01-21 13:40:00+00:00 521 PM1 6.90 2021-01-21 13:40:00]
[ Timestamp Location Name Value Date Time
670170 2023-05-19 08:20:01+00:00 521 PM1 8.67 2023-05-19 08:20:01
670171 2023-05-19 08:20:01+00:00 521 PM2.5 12.33 2023-05-19 08:20:01
670172 2023-05-19 08:20:01+00:00 521 PM10 19.85 2023-05-19 08:20:01, Timestamp Location Name Value Date Time
670177 2023-05-19 08:30:00+00:00 521 PM1 9.25 2023-05-19 08:30:00
670178 2023-05-19 08:30:00+00:00 521 PM2.5 12.79 2023-05-19 08:30:00
670179 2023-05-19 08:30:00+00:00 521 PM10 29.53 2023-05-19 08:30:00, Timestamp Location Name Value Date Time
670184 2023-05-19 08:40:00+00:00 521 PM1 8.97 2023-05-19 08:40:00
670185 2023-05-19 08:40:00+00:00 521 PM2.5 12.51 2023-05-19 08:40:00
670186 2023-05-19 08:40:00+00:00 521 PM10 21.14 2023-05-19 08:40:00, Timestamp Location Name Value Date Time
670191 2023-05-19 08:50:00+00:00 521 PM1 9.54 2023-05-19 08:50:00
670192 2023-05-19 08:50:00+00:00 521 PM2.5 13.44 2023-05-19 08:50:00
670193 2023-05-19 08:50:00+00:00 521 PM10 19.53 2023-05-19 08:50:00, Timestamp Location Name Value Date Time
670198 2023-05-19 09:00:00+00:00 521 PM1 8.15 2023-05-19 09:00:00
670199 2023-05-19 09:00:00+00:00 521 PM2.5 11.33 2023-05-19 09:00:00
670200 2023-05-19 09:00:00+00:00 521 PM10 26.28 2023-05-19 09:00:00]
It looks like the grouping is going according to plan, so now we want to use the info from each grouping to create a row where the different names here are different columns of the row.
for x in date_time_groups.groups:
current_group = date_time_groups.get_group(x)
location = current_group['Location'][current_group.index[0]]
new_row = {'Date': [x[0]], 'Time': [x[1]], 'Location': [location], 'PM10': [np.nan], 'PM2.5': [np.nan],
'PM1': [np.nan]}
for row in current_group.values:
new_row[row[2]] = [row[3]]
df_521_measurements = pd.concat([df_521_measurements, pd.DataFrame(new_row)])
print(df_521_measurements)
Date Time Location PM10 PM2.5 PM1
0 2021-01-21 13:00:00 521 22.03 10.54 7.29
0 2021-01-21 13:10:00 521 20.90 10.52 7.02
0 2021-01-21 13:20:00 521 21.30 10.93 6.66
0 2021-01-21 13:30:00 521 20.23 11.72 7.67
0 2021-01-21 13:40:00 521 19.25 11.37 6.90
.. ... ... ... ... ... ...
0 2023-05-19 08:20:01 521 19.85 12.33 8.67
0 2023-05-19 08:30:00 521 29.53 12.79 9.25
0 2023-05-19 08:40:00 521 21.14 12.51 8.97
0 2023-05-19 08:50:00 521 19.53 13.44 9.54
0 2023-05-19 09:00:00 521 26.28 11.33 8.15
[111871 rows x 6 columns]
The measurements do all have an index of 0 now, so we do want to reset the index, as it is already in order.
df_521_measurements = df_521_measurements.reset_index(drop=True)
Now we want to visualize a single day at a single location, to see any quick patterns.
df_521_measurements.Date[0]
datetime.date(2021, 1, 21)
df_lcd = df_521_measurements.loc[df_521_measurements['Date'] == pd.to_datetime('2023-02-20')]
print(df_lcd)
Date Time Location PM10 PM2.5 PM1
99235 2023-02-20 00:00:00 521 20.17 15.49 13.95
99236 2023-02-20 00:10:00 521 20.61 15.38 13.11
99237 2023-02-20 00:20:00 521 22.29 15.65 12.96
99238 2023-02-20 00:30:00 521 21.63 15.73 13.35
99239 2023-02-20 00:40:01 521 23.97 16.24 14.77
... ... ... ... ... ... ...
99373 2023-02-20 23:10:01 521 39.96 29.24 25.05
99374 2023-02-20 23:20:00 521 52.30 32.50 25.66
99375 2023-02-20 23:30:00 521 42.31 31.38 24.80
99376 2023-02-20 23:40:00 521 42.03 30.57 24.89
99377 2023-02-20 23:50:00 521 39.30 30.58 23.82
[143 rows x 6 columns]
fig, ax = plt.subplots()
fig.canvas.draw()
plt.scatter(df_lcd.index, df_lcd['PM10'], label='PM10', alpha=0.5)
plt.scatter(df_lcd.index, df_lcd['PM2.5'], label='PM2.5', alpha=0.5)
plt.scatter(df_lcd.index, df_lcd['PM2.5'], label='PM1', alpha=0.5)
ax.set_xticks(df_lcd.index[::6])
ax.set_xticklabels(df_lcd['Time'][::6], rotation=90)
fig.suptitle('20-2-2023 Fine dust particles')
ax.set_xlabel('Time')
ax.set_ylabel('Particles (μg/m³)')
plt.legend()
plt.show()
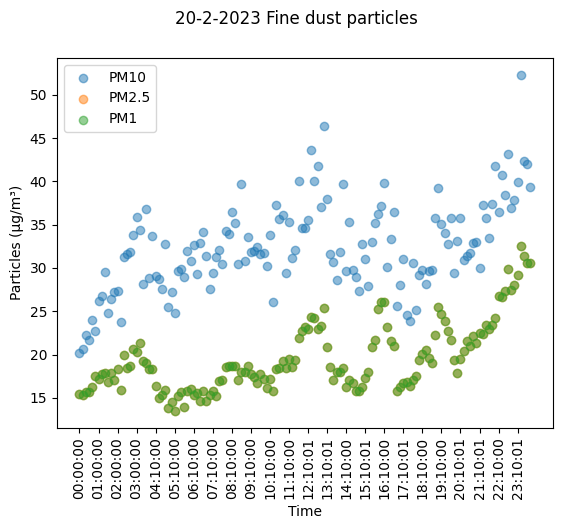
From here we can see that PM1 and PM2.5 are near identical, at least in comparison to PM10. If we look at the actual data we can see there are differences. We can also see that there is some data missing it seems, as after 3:00:00 comes 4:10:00, so somewhere along the lines some 10 minutes went missing it seems. Another thing we can observe is that the increases and decreases of the different particle matters are at similar moments, so there are probably some trends to observe there.
measurement_521_drop_unused
| Timestamp | Location | Name | Value | Date | Time | |
|---|---|---|---|---|---|---|
| 54 | 2021-01-21 13:00:00+00:00 | 521 | PM10 | 22.03 | 2021-01-21 | 13:00:00 |
| 55 | 2021-01-21 13:00:00+00:00 | 521 | PM2.5 | 10.54 | 2021-01-21 | 13:00:00 |
| 56 | 2021-01-21 13:00:00+00:00 | 521 | PM1 | 7.29 | 2021-01-21 | 13:00:00 |
| 59 | 2021-01-21 13:10:00+00:00 | 521 | PM10 | 20.90 | 2021-01-21 | 13:10:00 |
| 60 | 2021-01-21 13:10:00+00:00 | 521 | PM2.5 | 10.52 | 2021-01-21 | 13:10:00 |
| ... | ... | ... | ... | ... | ... | ... |
| 670192 | 2023-05-19 08:50:00+00:00 | 521 | PM2.5 | 13.44 | 2023-05-19 | 08:50:00 |
| 670193 | 2023-05-19 08:50:00+00:00 | 521 | PM10 | 19.53 | 2023-05-19 | 08:50:00 |
| 670198 | 2023-05-19 09:00:00+00:00 | 521 | PM1 | 8.15 | 2023-05-19 | 09:00:00 |
| 670199 | 2023-05-19 09:00:00+00:00 | 521 | PM2.5 | 11.33 | 2023-05-19 | 09:00:00 |
| 670200 | 2023-05-19 09:00:00+00:00 | 521 | PM10 | 26.28 | 2023-05-19 | 09:00:00 |
335613 rows × 6 columns
df_521_PM10 = measurement_521_drop_unused.loc[measurement_521_drop_unused['Name'] == "PM10"]
df_521_PM1 = measurement_521_drop_unused.loc[measurement_521_drop_unused['Name'] == "PM1"]
df_521_PM25 = measurement_521_drop_unused.loc[measurement_521_drop_unused['Name'] == "PM2.5"]
df_521_PM25 = df_521_PM25.set_index('Timestamp')
df_521_PM1 = df_521_PM1.set_index('Timestamp')
df_521_PM10 = df_521_PM10.set_index('Timestamp')
fig, axs = plt.subplots(3, sharex=True, figsize=(15, 10))
fig.suptitle('The different sizes of particles individual values on location 521')
axs[0].plot(df_521_PM10['Value'], 'tab:blue')
axs[0].title.set_text('PM10')
axs[1].plot(df_521_PM25['Value'], 'tab:orange')
axs[1].title.set_text('PM25')
axs[2].plot(df_521_PM1['Value'], 'tab:green')
axs[2].title.set_text('PM1')
plt.tight_layout()
plt.subplots_adjust(hspace=0.75)
plt.xticks(rotation=90)
plt.show()
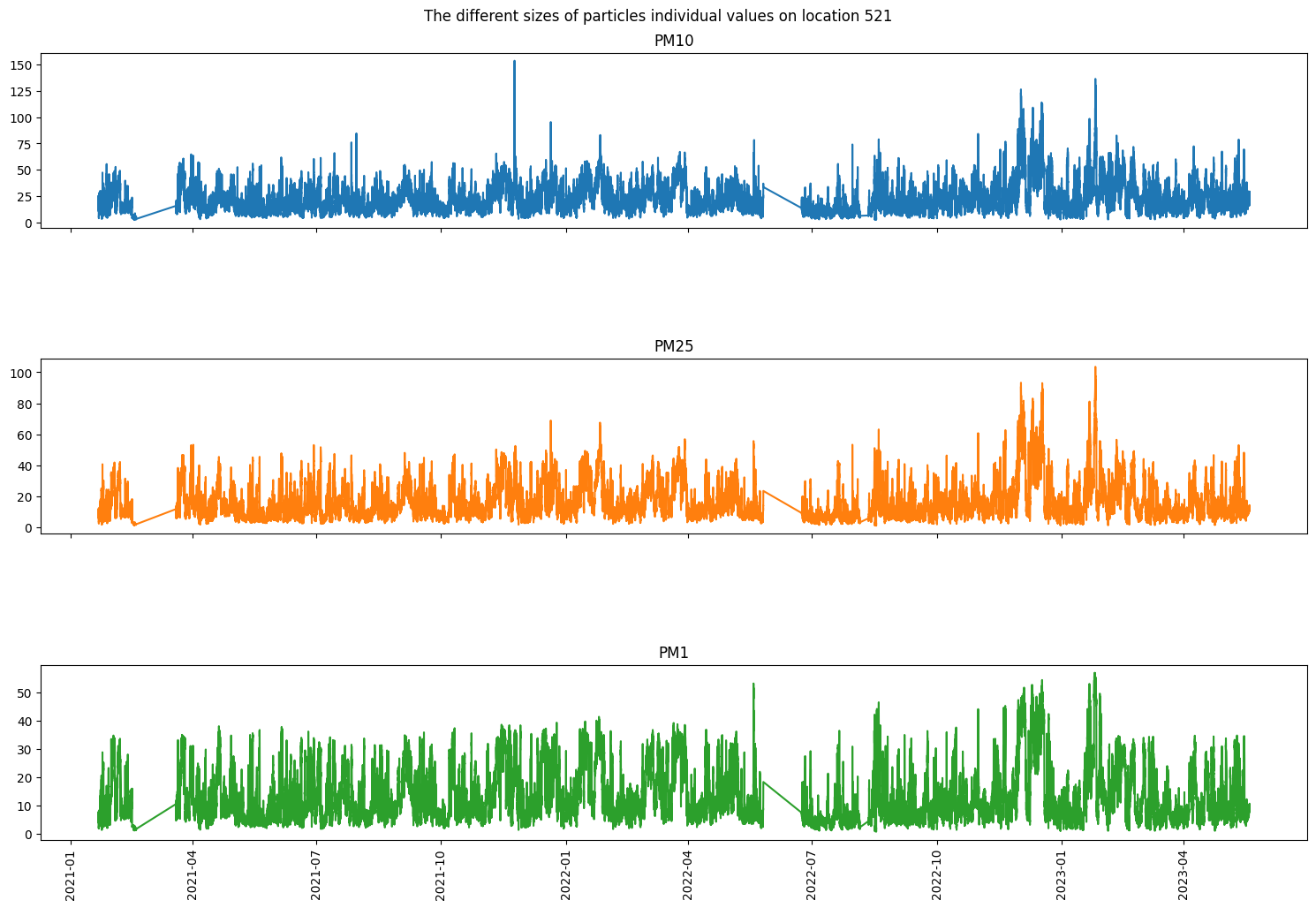
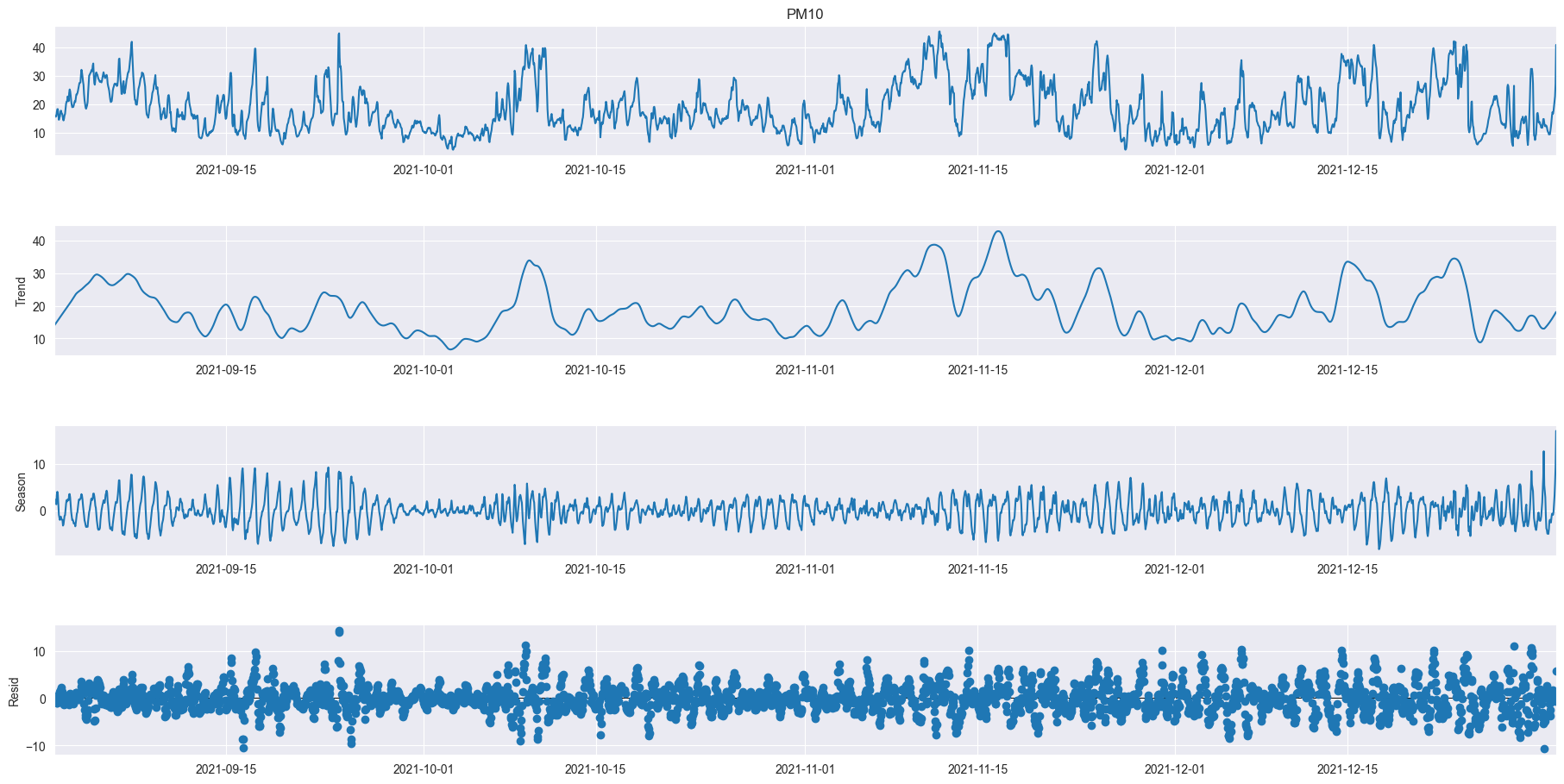
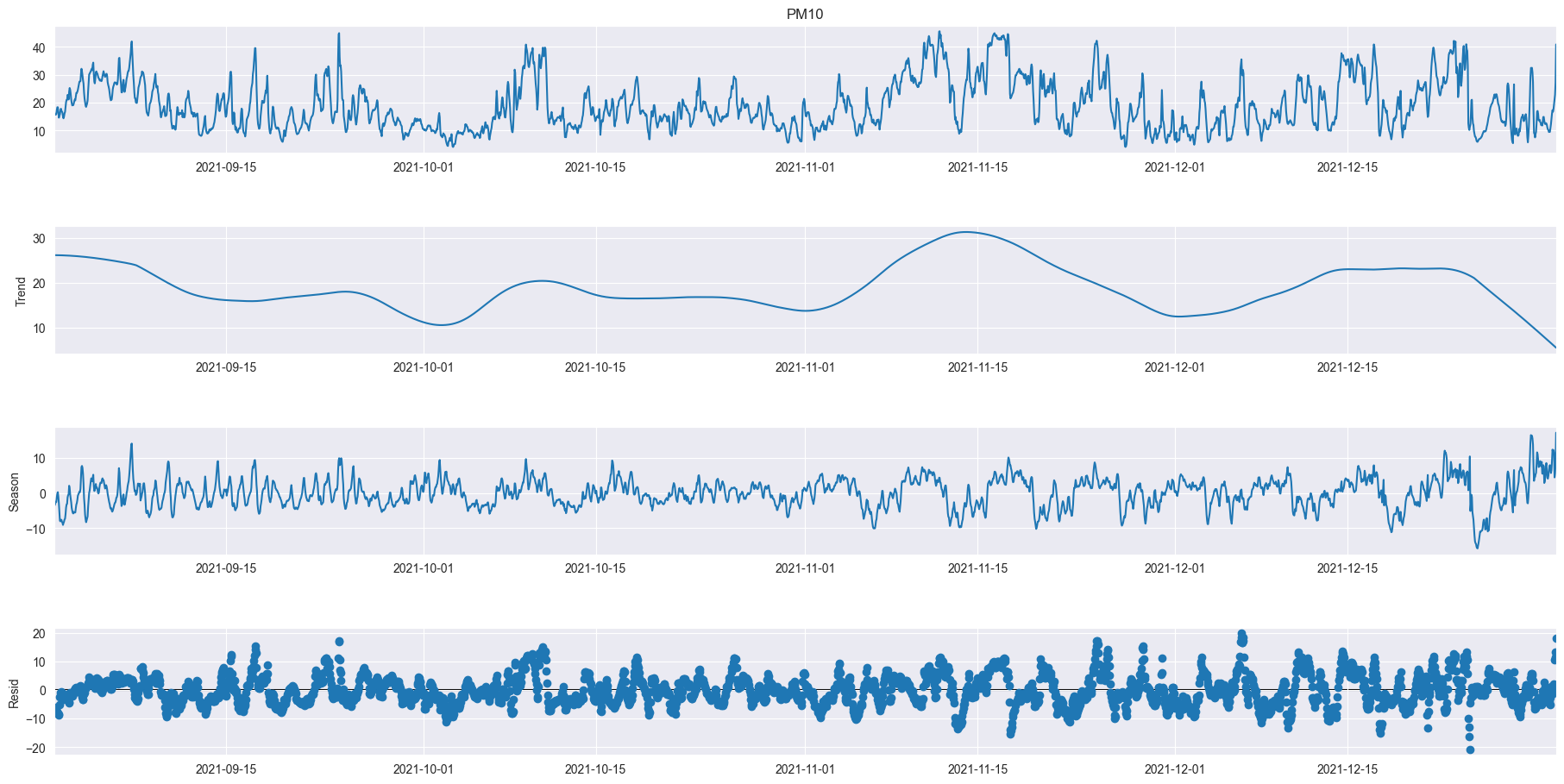
As we can see even over a longer period of time there are definitely relations between the different sizes of atoms but also individuality. We can also very clearly see two moments where the data was not recorded for a long period of time. In PM10 we can see a little outlier spike at the end of 2021, in PM1 there is an outlier halfway through 2022 and PM2.5 doesn’t seem to have any notable outliers.
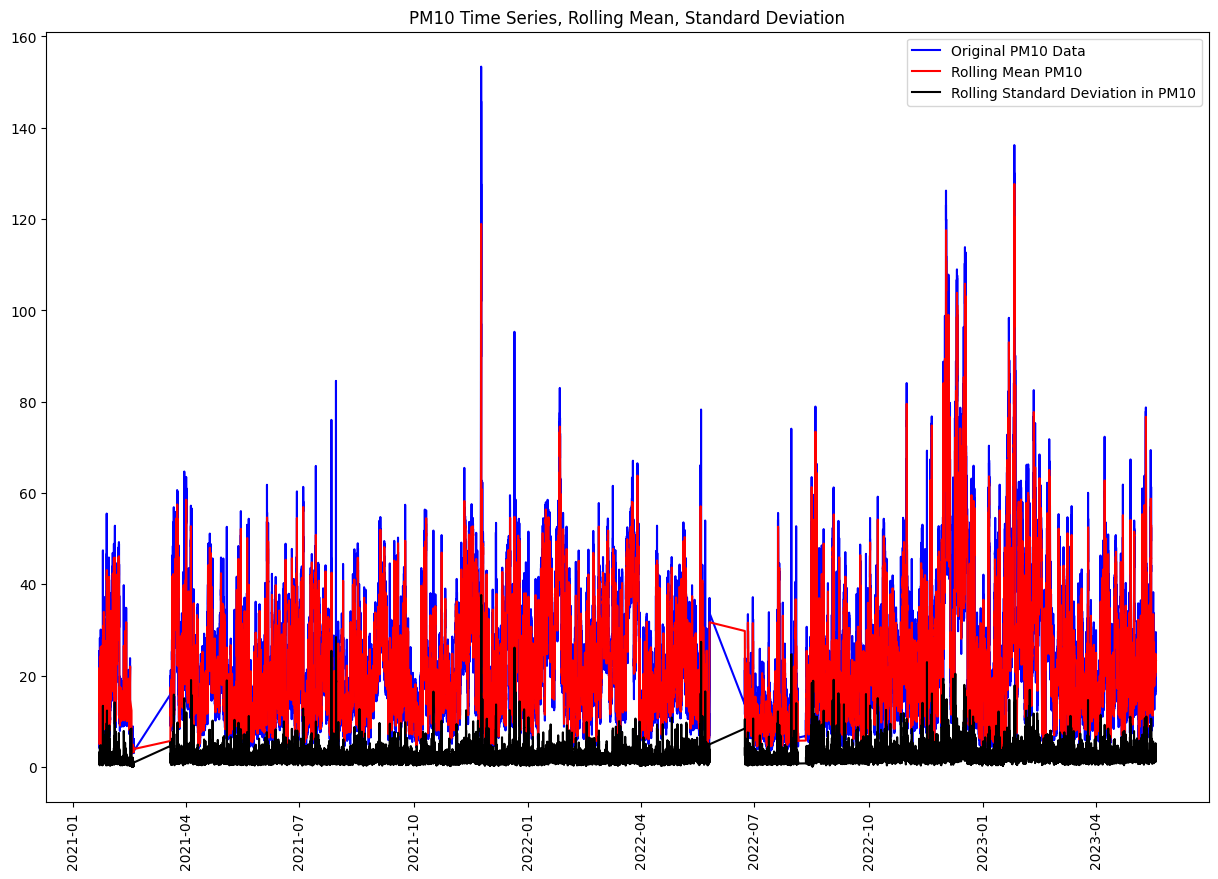
rolling_mean = df_521_PM10['Value'].rolling(7).mean()
rolling_std = df_521_PM10['Value'].rolling(7).std()
plt.figure(figsize=(15, 10))
plt.plot(df_521_PM10['Value'], color="blue", label="Original PM10 Data")
plt.plot(rolling_mean, color="red", label="Rolling Mean PM10")
plt.plot(rolling_std, color="black", label="Rolling Standard Deviation in PM10")
plt.title("PM10 Time Series, Rolling Mean, Standard Deviation")
plt.legend(loc="best")
plt.xticks(rotation=90)
plt.show()
from statsmodels.tsa.stattools import adfuller
adft = adfuller(df_521_PM10['Value'], autolag="AIC")
output_df = pd.DataFrame({"Values": [adft[0], adft[1], adft[2], adft[3], adft[4]['1%'], adft[4]['5%'], adft[4]['10%']],
"Metric": ["Test Statistics", "p-value", "No. of lags used", "Number of observations used",
"critical value (1%)", "critical value (5%)", "critical value (10%)"]})
print(output_df)
Values Metric
0 -1.762838e+01 Test Statistics
1 3.808919e-30 p-value
2 6.500000e+01 No. of lags used
3 1.118050e+05 Number of observations used
4 -3.430408e+00 critical value (1%)
5 -2.861566e+00 critical value (5%)
6 -2.566784e+00 critical value (10%)
The most important part of this is the p-value, or the probability that a particular statistical measure, such as the mean or standard deviation, of an assumed probability distribution will be greater than or equal to (or less than or equal to in some instances) observed results. We have a very small p-value so that is good.
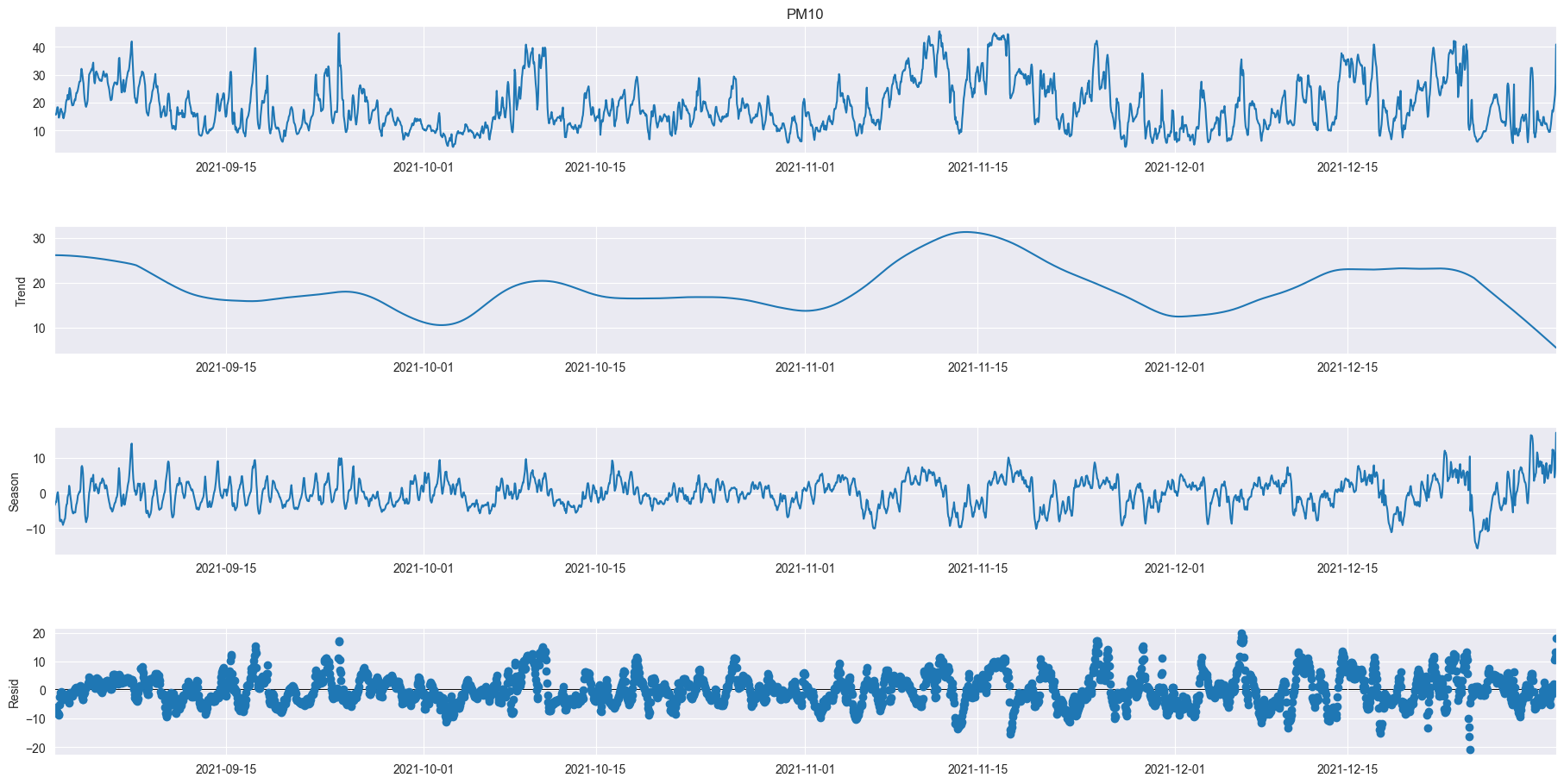
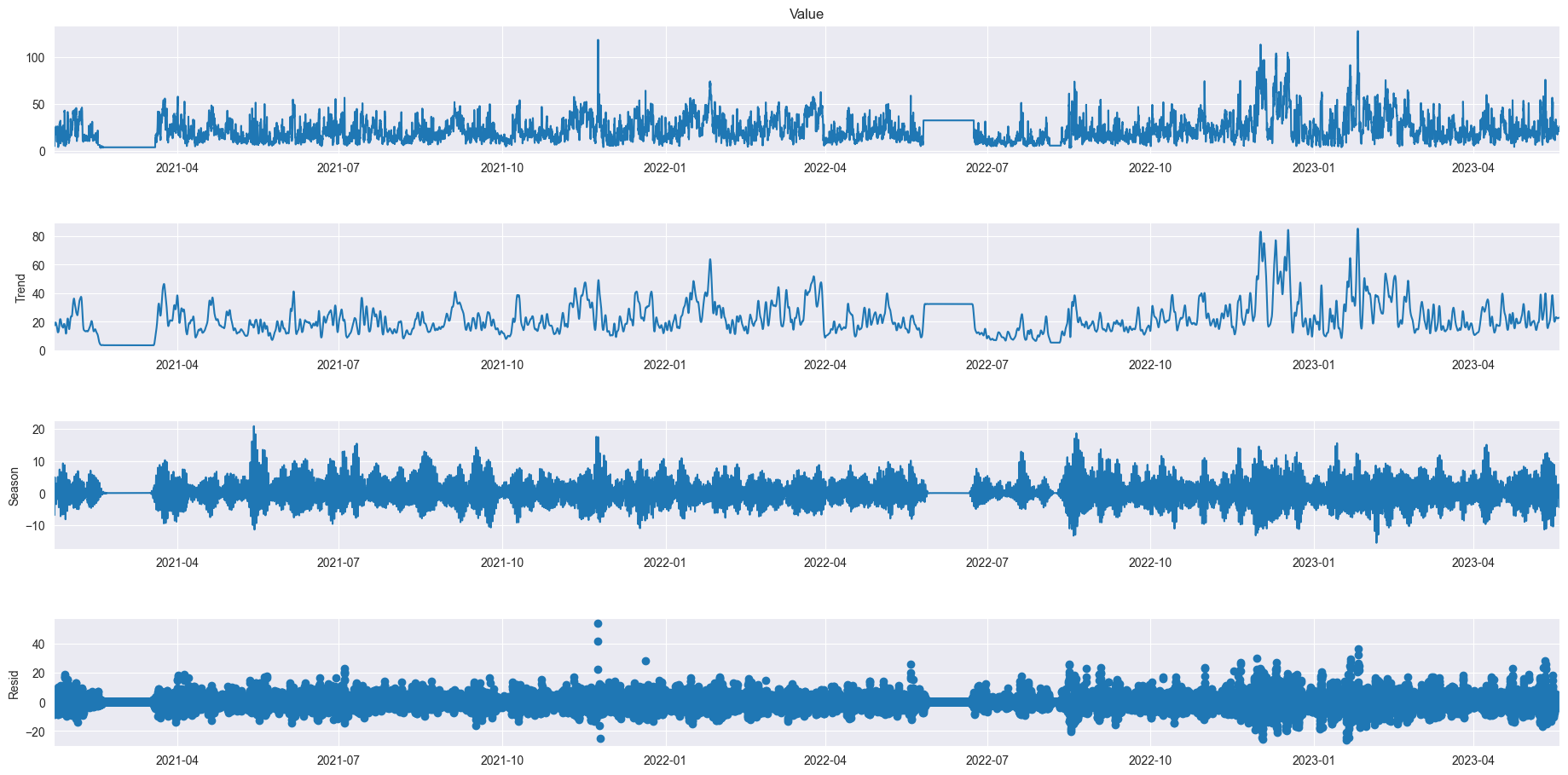
from statsmodels.tsa.seasonal import STL
# plt.figure(figsize=(60, 20))
data = df_521_PM10.resample('10min').mean().ffill()
res = STL(data['Value'], period=144).fit()
res.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
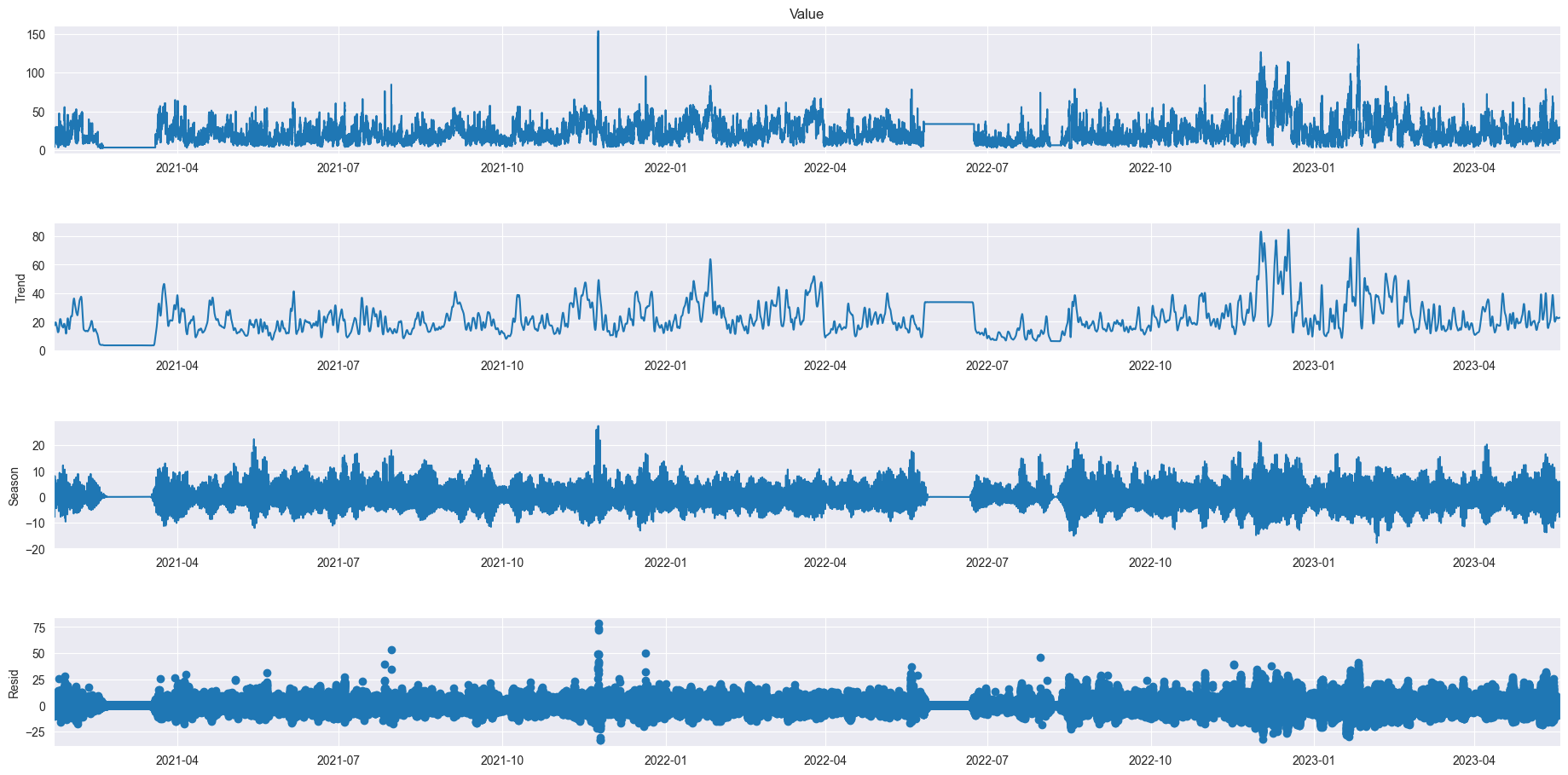
# plt.figure(figsize=(30, 20))
res2 = STL(data['Value'], period=1008).fit()
res2.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
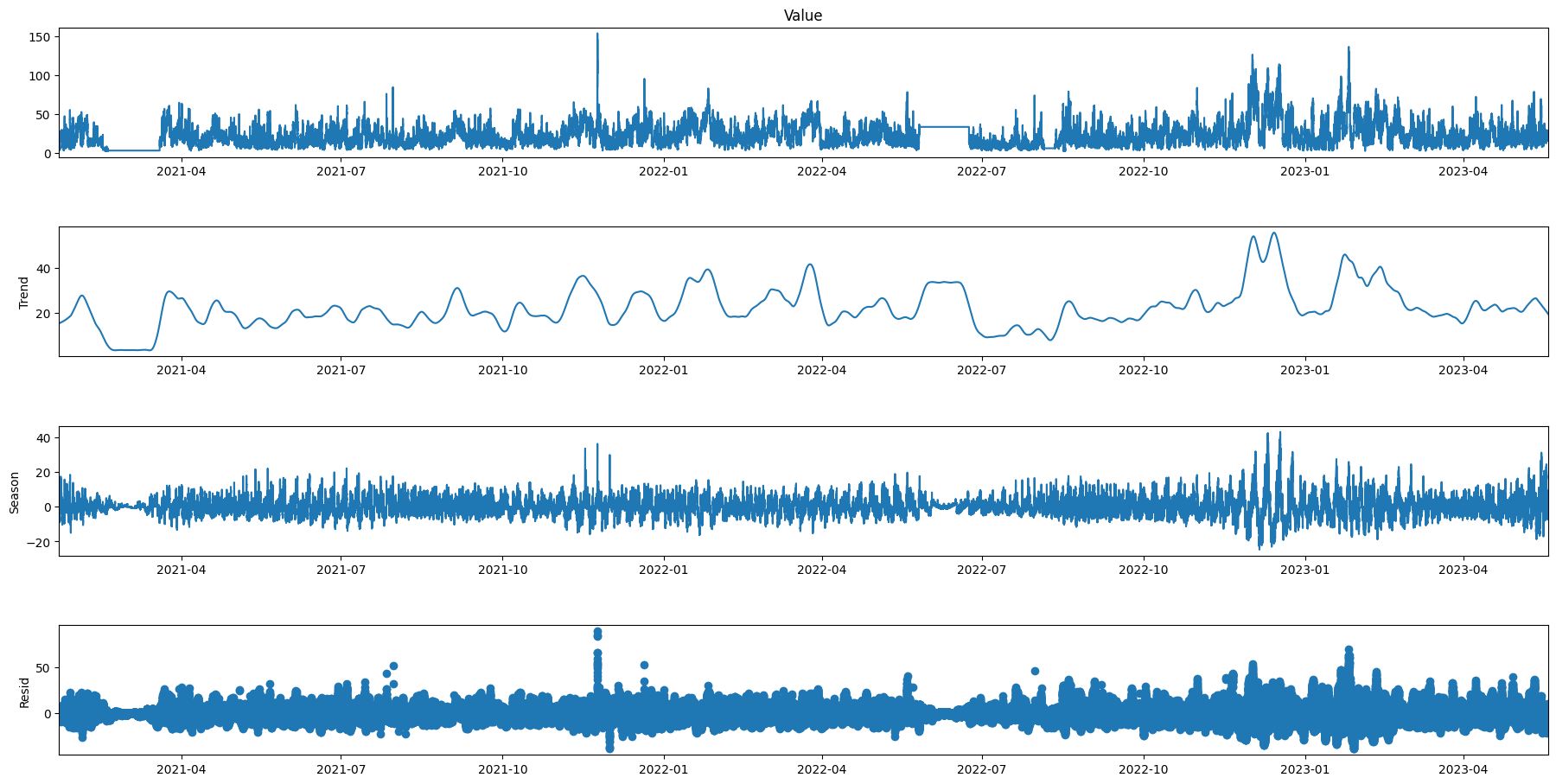
# plt.figure(figsize=(30, 20))
res3 = STL(data['Value'], period=30240).fit()
res3.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
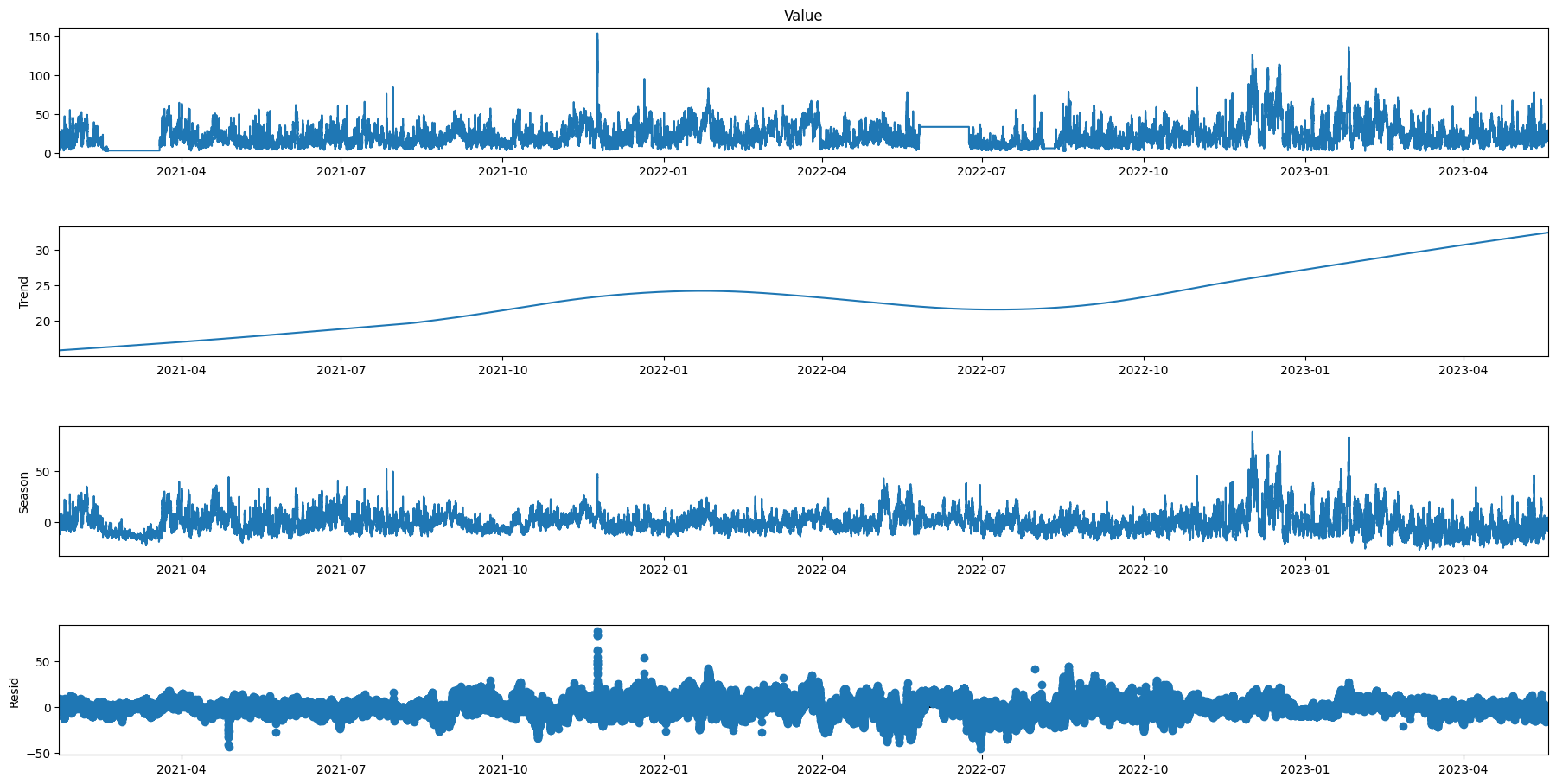
Test with weekly seasonality over 1 months
subset1 = data.loc['2021-04-01':'2021-04-30']
res4 = STL(subset1['Value'], period=1008).fit()
res4.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
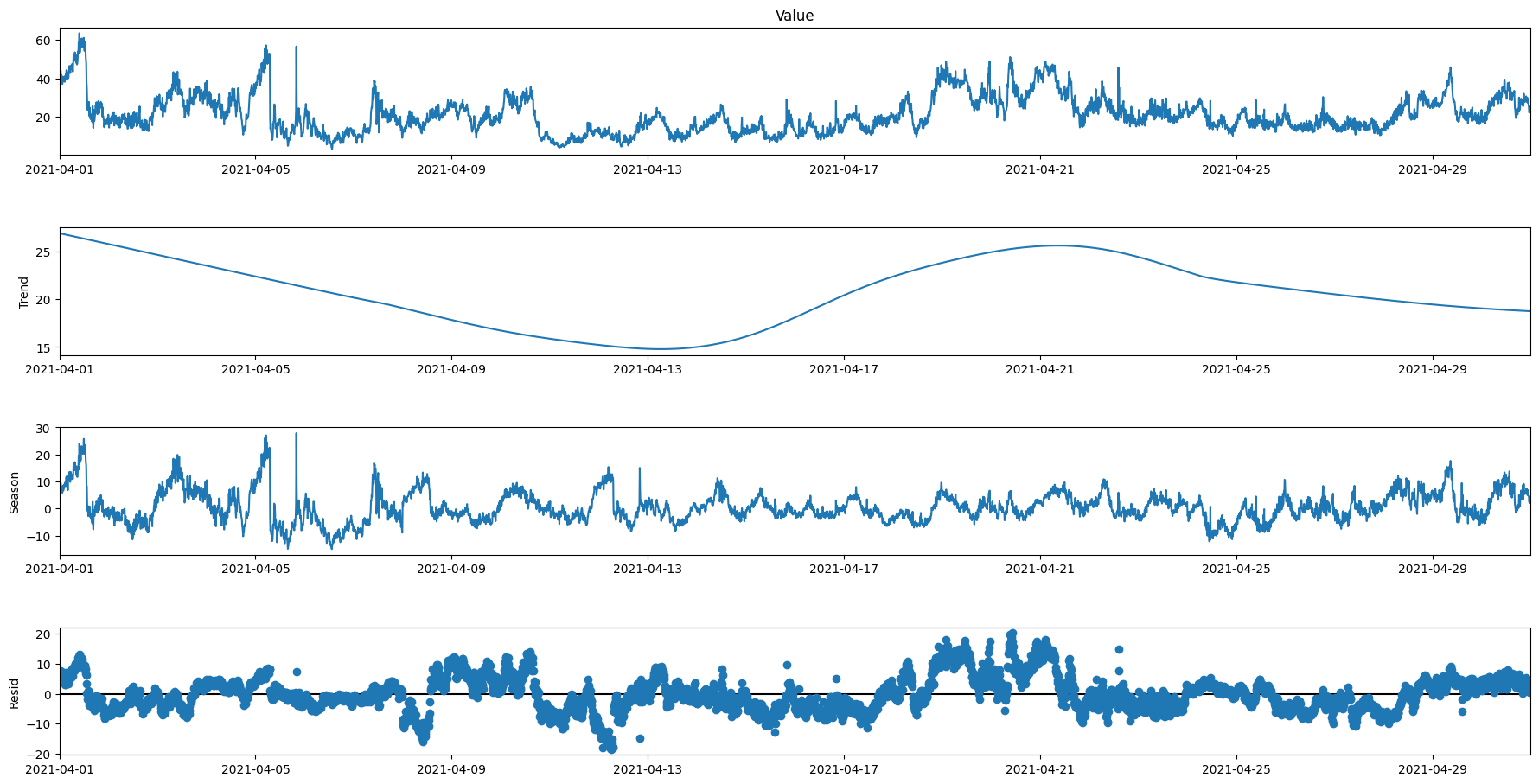
Combining all the data
Now we have all the data and we know what a single location looks like and how we need to alter the data so we can use it, we can combine all the data into a single dataset to work from.
location_data = pd.DataFrame(columns=['location_id', 'Lat', 'Lon'])
measurement_full = pd.DataFrame(columns=['Date', 'Time', 'Location', 'PM10', 'PM2.5', 'PM1'])
for dataset in file_list:
current_df = pd.read_csv(file_path + dataset)
#get location
lon = current_df.loc[current_df['Name'] == 'Lon']
lat = current_df.loc[current_df['Name'] == 'Lat']
loc_data = {'location_id': [current_df['Location'][0]],
'Lat': [lat.groupby('Value')['Value'].count().sort_values(ascending=False).head(1).index.values[0]],
'Lon': [lon.groupby('Value')['Value'].count().sort_values(ascending=False).head(1).index.values[0]]}
location_data = pd.concat([location_data, pd.DataFrame(loc_data)])
#get measurements
df_measurements_data = current_df.loc[:, ['Timestamp', 'Location', 'Name', 'Value']]
df_measurements_drop_unused = df_measurements_data[
(df_measurements_data.Name != 'Lat') & (df_measurements_data.Name != 'Lon') & (
df_measurements_data.Name != 'NO2') & (df_measurements_data.Name != 'UFP')].copy()
df_measurements_drop_unused['Timestamp'] = pd.to_datetime(df_measurements_drop_unused['Timestamp'])
df_measurements_drop_unused['Date'] = [d.date() for d in df_measurements_drop_unused['Timestamp']]
df_measurements_drop_unused['Time'] = [d.time() for d in df_measurements_drop_unused['Timestamp']]
df_measurements = pd.DataFrame(columns=['Date', 'Time', 'Location', 'PM10', 'PM2.5', 'PM1'])
date_time_groups = df_measurements_drop_unused.groupby(['Date', 'Time'])
for x in date_time_groups.groups:
current_group = date_time_groups.get_group(x)
location = current_group['Location'][current_group.index[0]]
new_row = {'Date': [x[0]], 'Time': [x[1]], 'Location': [location], 'PM10': [np.nan], 'PM2.5': [np.nan],
'PM1': [np.nan]}
for row in current_group.values:
new_row[row[2]] = [row[3]]
df_measurements = pd.concat([df_measurements, pd.DataFrame(new_row)])
measurement_full = pd.concat([measurement_full, df_measurements])
print(dataset)
Everything is combined, so let’s take a look at whether everything is like we want it to.
It seems like only the index is still a problem, which was to be expected. So if we reset the index, we can save the data into a single file so we can later just load that one to continue preparing and analysing our data.
measurement_full = measurement_full.reset_index(drop=True)
measurement_full.to_csv(
"C:/Users/Public/Documents/Github/Internship-S5/cdt-a-i-r-zoom-lens-back-end/csv_files/full_measurement.csv",
index=False)
len(measurement_full)
We also need to save the location data that we seperated from our raw data.
location_data.to_csv(
"C:/Users/Public/Documents/Github/Internship-S5/cdt-a-i-r-zoom-lens-back-end/csv_files/location_data.csv",
index=False)
Seeing as we now saved them in a seperate directory than the raw directory, we also want a new file path to work with.
Analysing the combined data
Now let’s get our dataframes again and start analysing them.
df_file_path = "C:/Users/Public/Documents/Github/Internship-S5/cdt-a-i-r-zoom-lens-back-end/csv_files/"
loc_df = pd.read_csv(df_file_path + 'location_data.csv')
measurement_df = pd.read_csv(df_file_path + 'full_measurement.csv')
print(loc_df.sample(10))
print(measurement_df.sample(10))
location_id Lat Lon
18 542 51.5108 5.3606
20 544 51.4379 5.3582
26 553 51.4915 5.4392
41 570 51.5835 5.5480
31 560 51.4414 5.4714
0 521 51.3468 5.3887
15 537 52.6558 4.7969
29 557 51.5696 5.3113
9 531 51.4063 5.4574
17 540 51.4797 5.4751
Date Time Location PM10 PM2.5 PM1
3883319 2023-02-06 06:50:01 560 11.67 8.65 7.86
4226370 2022-11-18 04:46:00 563 7.50 3.81 2.93
1319576 2022-05-27 07:00:01 532 11.91 7.19 4.55
5072896 2022-01-11 05:40:00 571 31.12 26.72 24.59
1790736 2022-08-17 08:50:00 536 23.60 21.00 19.39
895283 2021-05-26 03:10:00 529 15.61 7.72 5.06
4916557 2022-11-29 08:10:00 569 30.32 30.17 26.16
5008998 2022-08-22 21:40:00 570 7.80 5.04 3.45
1865333 2021-06-24 01:30:00 537 9.88 7.38 6.62
4726573 2023-05-08 00:40:00 567 15.60 11.00 10.30
When we were scraping the data, we noticed that location 575 took much longer than the rest, as it was getting data from 2015. However, let’s see how much of that data is still relevant to us.
longest_location = measurement_df[(measurement_df.Location == 575)]
print(longest_location.head(10))
Date Time Location PM10 PM2.5 PM1
5390315 2019-04-18 11:50:00 575 19.54 11.46 9.90
5390316 2019-04-18 12:00:00 575 19.95 10.91 9.86
5390317 2019-04-18 12:10:00 575 28.21 11.53 10.39
5390318 2019-04-18 12:20:00 575 14.92 10.62 9.07
5390319 2019-04-18 12:40:00 575 18.03 9.98 8.84
5390320 2019-04-18 12:50:00 575 13.04 8.44 6.86
5390321 2019-04-18 13:20:00 575 14.63 7.20 6.47
5390322 2019-04-18 13:30:00 575 14.60 6.50 5.67
5390323 2019-04-18 13:40:00 575 12.77 6.90 5.94
5390324 2019-04-18 13:50:00 575 16.92 6.87 6.03
It looks like though it started in 2015, the data we need didn’t get recorded until April 2019. Let’s see what that is like with the other locations, so we know the distribution of when our actual data got collected.
starting_dates = []
for location in measurement_df.Location.unique():
location_df = measurement_df[(measurement_df.Location == location)]['Date']
starting_dates.append(location_df.head(1).values[0])
print(starting_dates)
['2021-01-21', '2020-10-19', '2020-12-02', '2020-12-02', '2020-09-14', '2020-09-14', '2021-01-21', '2020-10-19', '2019-05-16', '2021-03-30', '2020-12-02', '2020-10-19', '2020-12-02', '2020-10-19', '2020-09-14', '2020-10-19', '2020-12-02', '2020-10-19', '2020-12-02', '2020-09-14', '2020-09-14', '2020-12-02', '2020-10-19', '2021-01-20', '2020-11-09', '2020-10-19', '2020-12-02', '2021-01-21', '2020-12-02', '2021-01-22', '2020-10-19', '2020-10-19', '2021-01-21', '2020-09-14', '2021-01-22', '2020-09-14', '2021-01-21', '2020-09-14', '2021-01-21', '2021-01-21', '2021-05-01', '2021-03-30', '2021-03-30', '2021-03-26', '2021-05-01', '2019-04-18', '2019-04-18', '2021-05-01', '2021-05-01']
Now we have the first date of relevant data of each location, let’s see how it is distributed.
from collections import Counter, OrderedDict
c = Counter(starting_dates)
ordered_data = OrderedDict(sorted(c.items(), key=lambda x: datetime.strptime(x[0], '%Y-%m-%d'), reverse=False))
plt.bar(ordered_data.keys(), ordered_data.values())
plt.suptitle("Amount of locations that started recording relevant data on that date")
plt.xticks(rotation=90)
plt.xlabel("Start date")
plt.ylabel("Amount of locations")
plt.show()
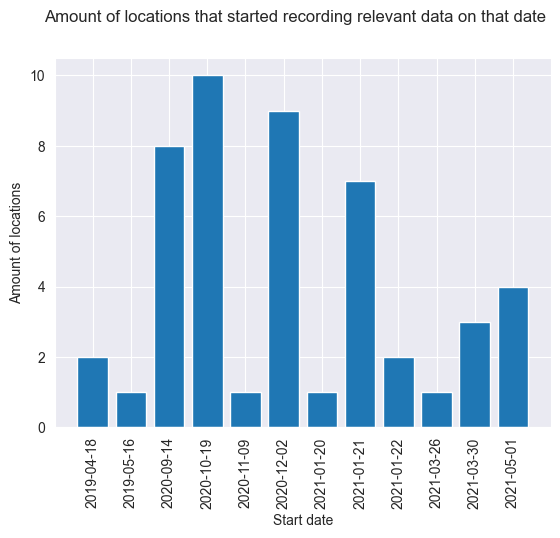
As we can see, there were two locations that started recording in April 2019, which were the first. So the earliest records are from April 18th 2019. Most locations started recording at the end of 2020, somewhere between September 14th and December 2nd. There were also still quite some locations that started recording at the start of 2021, and the latest a location started recording was May 2021. Something of note here, is that this means that we have 2 sensors that were placed before covid, while the rest was placed during. This means that outliers in observations don’t have a ‘base level’ from before covid to compare it with. This needs to be taken into account when analysing the results.
Something else that we need to verify, is whether everything is in the same unit. We know at location 521 it was, but we need to verify whether this is the case with all the locations.
units = []
for file in file_list:
df_temp = pd.read_csv(file_path + file)
unique_units = df_temp.groupby('Name')['Unit'].unique()
units.append(unique_units)
unit_df = pd.DataFrame()
for unit in range(len(units)):
for row in range(len(units[unit])):
units[unit][row] = units[unit][row][0]
unit_df = pd.concat([unit_df, units[unit].to_frame().T], ignore_index=True)
unique_unit = unit_df.drop_duplicates()
print(unique_unit)
Name Lat Lon NO2 PM1 PM10 PM2.5 UFP
0 ° ° μg/m³\r\n μg/m³ μg/m³ μg/m³ counts/cm³\r\n
6 ° ° μg/m³\r\n μg/m³ μg/m³ μg/m³ NaN
It looks like the only place where there is a difference in the unit values, is in a location where the UFP unit is NaN. However, seeing as we are not doing anything with the UFP unit, this doesn’t matter, so now we have verified that the same unit of measurement is used in every single location. Now we have verified everything we needed, we can take a definitive look at the data quality.
Data Quality
Usually you would check the data quality before you prepare the data, so you can adjust accordingly. However, in our case preparing the data and collecting it was intertwined, adjusting the way of collecting so there was less need for preparing it. That is why the data quality is checked after the initial preparation of the data, as the data is not yet completely prepared, but enough that we can work with it.
Validity - Check whether there is always a number when we expect it etc Accuracy - We cannot definitevely say it is accurate, for example the location keeps jumping so it is prone to mistakes, but we don't need 100% accurate + this is why we trust this source and sample a few from samenmeten and explain some differences Consistency - The different units are the same Completeness - Missing data, how much do we have etc Uniqueness - It is not unique, as samenmeten enzo het ook heeft, but this is the difference between those things Timeliness - Check the time it takes to make a request to the api for 1 location for right now
Validity
Let’s start by looking at info to see whether the type matches.
measurement_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 5739479 entries, 0 to 5739478
Data columns (total 6 columns):
# Column Dtype
--- ------ -----
0 Date object
1 Time object
2 Location int64
3 PM10 float64
4 PM2.5 float64
5 PM1 float64
dtypes: float64(3), int64(1), object(2)
memory usage: 262.7+ MB
It looks like the type is matching with what we are expecting, the date and time are objects and location, location is an int as it an ID and PM10, PM2.5 and PM1 are all floats as they are measurements. However, let’s check whether they are all positive, as a negative amount of fine dust does not exist.
measurement_df.describe()
| Location | PM10 | PM2.5 | PM1 | |
|---|---|---|---|---|
| count | 5.739479e+06 | 5.735059e+06 | 5.739478e+06 | 5.739479e+06 |
| mean | 5.485570e+02 | 1.835883e+01 | 1.273283e+01 | 1.031460e+01 |
| std | 1.692974e+01 | 1.443371e+01 | 1.396131e+01 | 1.273105e+01 |
| min | 5.210000e+02 | -0.000000e+00 | -3.200000e+00 | -3.590000e+00 |
| 25% | 5.330000e+02 | 9.990000e+00 | 5.500000e+00 | 4.120000e+00 |
| 50% | 5.470000e+02 | 1.518000e+01 | 9.330000e+00 | 7.140000e+00 |
| 75% | 5.640000e+02 | 2.360000e+01 | 1.652000e+01 | 1.352000e+01 |
| max | 5.770000e+02 | 1.196890e+03 | 1.358270e+03 | 1.355330e+03 |
As we can see, there are some negative values. Not a lot, as they do not reach past the 25% quartile, however they do exist. Let’s see how often they occur.
negative_percentages = pd.DataFrame()
for location in measurement_df.Location.unique():
loc_data = measurement_df[(measurement_df.Location == location)]
neg_data = loc_data[(loc_data.PM10 < 0) | (loc_data['PM2.5'] < 0) | (loc_data.PM1 < 0)]
neg_df_data = {'loc_id': [location], 'neg_amounts': [len(neg_data)], 'total_amounts': [len(loc_data)],
'percentage_negative': [(len(neg_data) / len(loc_data)) * 100]}
negative_percentages = pd.concat([negative_percentages, pd.DataFrame(neg_df_data)])
print(negative_percentages.sort_values('percentage_negative', ascending=False))
loc_id neg_amounts total_amounts percentage_negative
0 566 5662 120626 4.693847
0 537 3388 130891 2.588413
0 573 805 43670 1.843371
0 544 2304 127384 1.808704
0 553 2061 115215 1.788830
0 560 1689 118812 1.421574
0 572 672 74668 0.899984
0 562 1037 130176 0.796614
0 540 856 128359 0.666880
0 551 691 118343 0.583896
0 536 640 130430 0.490685
0 522 546 117745 0.463714
0 526 633 139222 0.454670
0 527 546 134782 0.405099
0 568 423 109085 0.387771
0 558 425 118283 0.359308
0 546 424 121509 0.348945
0 529 369 113568 0.324915
0 569 327 104493 0.312940
0 524 363 123710 0.293428
0 570 287 98287 0.292002
0 530 528 183580 0.287613
0 535 291 106338 0.273656
0 575 338 147377 0.229344
0 574 251 136859 0.183400
0 545 171 125742 0.135993
0 525 162 121321 0.133530
0 556 98 112554 0.087069
0 571 73 95844 0.076165
0 533 100 132893 0.075249
0 549 61 116901 0.052181
0 528 54 116834 0.046219
0 534 55 124645 0.044125
0 543 58 133510 0.043442
0 567 46 110198 0.041743
0 576 8 102129 0.007833
0 577 2 99658 0.002007
0 547 1 112618 0.000888
0 565 0 118270 0.000000
0 564 0 125884 0.000000
0 563 0 114562 0.000000
0 561 0 114681 0.000000
0 557 0 116113 0.000000
0 554 0 116303 0.000000
0 542 0 125188 0.000000
0 539 0 125269 0.000000
0 532 0 78550 0.000000
0 531 0 94529 0.000000
0 521 0 111871 0.000000
plt.hist(negative_percentages.percentage_negative)
plt.suptitle("Distribution of percentage of negative values")
plt.xlabel("% of negative values")
plt.ylabel("Distribution")
plt.show()
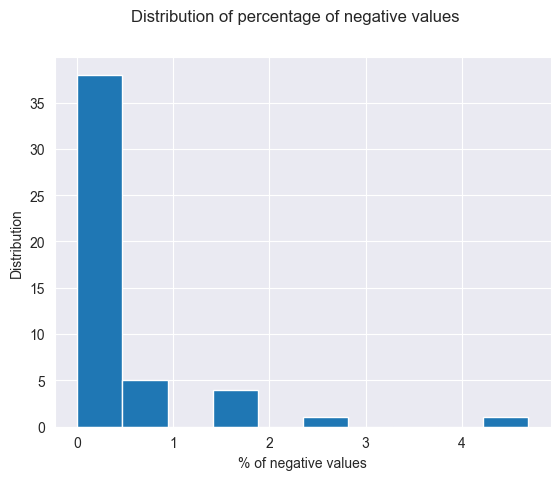
As we can see there are a few negative values, but most location have less than 1% or even no negative values. There are a few that are a bit higher than 1% and one is even just over 5%. It is not extremely concerning however it is a little bit damaging to the validity. However, the combination of correct types and only a few negative values means the *Validity of the data is decent, it is quite good but there is the issue with negative values.
Accuracy
We cannot definitively say the data is accurate. It is not something that you can classify, such as whether an animal is a dog. It is a measurement and measurements can be wrong and especially with something like fine dust, where the difference between the sensor and a meter away from the sensor can be very different, it is constantly changing based on the wind etc, we can only determine how much we trust we put in the idea that the data is accurate. For this we will look at a few aspects:
- Who is collecting it
- Who is using it
- Samenmeten
The data is being collected by ODZOB, the Omgevingsdienst Zuidoost-Brabant (Environment Service South-East Brabant). They work on executing the ecology and construction tasks mandated by law such as preventing noise disturbance, limiting health risks and tackling soil pollution {SOURCE, https://odzob.nl/expertises}. This means that the results of this organisation are trusted to be accurate enough for tasks mandated by law. This is also shown as they work with multiple municipalities. It is used to inform those who are making the local laws. Samenmeten, the portal of the RIVM, which is the national institute for public health and the environment, is also sharing the data of the sensors on their dataportal. So if local lawmakers are using the data, and the national institutes are sharing the data, and we know it is collected by experts, we can say that the Accuracy is acceptable.
Consistency
We know from checking the units just now, that the units used are consistent. So there is definetely a shared understanding of definition between the different locations. So we know the Consistency of the data is good.
Completeness
We know from the .info() that there aren’t any NaN values. However, this doesn’t mean that there isn’t any missing data. It just means that all the times that data was sent through, all the data was present. However when we were previously visualizing the single location data, we saw that there was a jump of 10 minutes, so that means that somewhere along the lines a measurement wasn’t collected once. Which isn’t that bad, however this was only once on the last collected day on one location. So let’s see how much missing timestamps there are overall. (Is there any missing data, how much do we have etc)
measurement_df.head()
| Date | Time | Location | PM10 | PM2.5 | PM1 | |
|---|---|---|---|---|---|---|
| 0 | 2021-01-21 | 13:00:00 | 521 | 22.03 | 10.54 | 7.29 |
| 1 | 2021-01-21 | 13:10:00 | 521 | 20.90 | 10.52 | 7.02 |
| 2 | 2021-01-21 | 13:20:00 | 521 | 21.30 | 10.93 | 6.66 |
| 3 | 2021-01-21 | 13:30:00 | 521 | 20.23 | 11.72 | 7.67 |
| 4 | 2021-01-21 | 13:40:00 | 521 | 19.25 | 11.37 | 6.90 |
import time
sets_of_10 = pd.DataFrame()
fmt = '%Y-%m-%d %H:%M:%S'
for location in measurement_df.Location.unique():
loc_data = measurement_df[(measurement_df.Location == location)]
d1 = datetime.strptime(loc_data.head(1).Date.values[0] + ' ' + loc_data.head(1).Time.values[0], fmt)
d2 = datetime.strptime(loc_data.tail(1).Date.values[0] + ' ' + loc_data.tail(1).Time.values[0], fmt)
d1_ts = time.mktime(d1.timetuple())
d2_ts = time.mktime(d2.timetuple())
supposed_sets = int(int(d2_ts - d1_ts) / 600)
sets_data = {'loc_id': [location], 'supposed_sets': [supposed_sets], 'actual_sets': [len(loc_data)],
'percentage_missing': [100 - (
(len(loc_data) / supposed_sets) * 100)]}
sets_of_10 = pd.concat([sets_of_10, pd.DataFrame(sets_data)])
print(sets_of_10)
loc_id supposed_sets actual_sets percentage_missing
0 521 122082 111871 8.364050
0 522 130448 117745 9.737980
0 524 129296 123710 4.320319
0 525 129296 121321 6.168018
0 526 140660 139222 1.022323
0 527 140661 134782 4.179552
0 528 122085 116834 4.301102
0 529 135641 113568 16.273103
0 530 210787 183580 12.907342
0 531 112308 94529 15.830573
0 532 94121 78550 16.543598
0 533 135639 132893 2.024491
0 534 129295 124645 3.596427
0 535 135640 106338 21.602772
0 536 140660 130430 7.272857
0 537 135641 130891 3.501891
0 539 129296 125269 3.114559
0 540 135641 128359 5.368583
0 542 129294 125188 3.175708
0 543 140659 133510 5.082504
0 544 140659 127384 9.437718
0 545 129295 125742 2.747979
0 546 135640 121509 10.418018
0 547 122214 112618 7.851801
0 549 131090 116901 10.823861
0 551 135635 118343 12.748922
0 553 129281 115215 10.880176
0 554 122083 116303 4.734484
0 556 129296 112554 12.948583
0 557 121926 116113 4.767646
0 558 135630 118283 12.789943
0 560 135641 118812 12.407016
0 561 122084 114681 6.063858
0 562 140661 130176 7.454092
0 563 121923 114562 6.037417
0 564 140661 125884 10.505400
0 565 122085 118270 3.124872
0 566 140662 120626 14.244074
0 567 122086 110198 9.737398
0 568 122086 109085 10.649051
0 569 107770 104493 3.040735
0 570 112312 98287 12.487535
0 571 112312 95844 14.662725
0 572 112863 74668 33.841915
0 573 49251 43670 11.331750
0 574 214835 136859 36.295762
0 575 214838 147377 31.400869
0 576 107775 102129 5.238692
0 577 107777 99658 7.533147
As we can see, there is definitely some missing timestamps. Now let’s see how much.
plt.hist(sets_of_10.percentage_missing, bins=10)
plt.suptitle('Distribution of Percentage of Missing Timestamps')
plt.xlabel('Percentage missing')
plt.ylabel('Amount of locations')
plt.show()
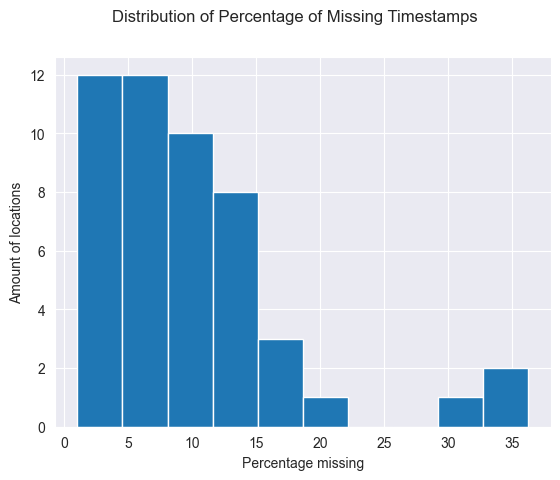
As we can see, most locations have around 10% of missing data or under. There are a few with more, however for now it looks like, with years of data every there is usually a max of 20% data loss over 45 locations. The 4 remaining locations, do have a larger amount of missing data, between 20 to 40%. However, it might be because at the start the data collection didn’t go as well. Let’s see which locations have this much missing data and whether they are the locations that have been around the longest.
highest_missing = sets_of_10.sort_values('percentage_missing', ascending=False)[:4]
for location in highest_missing.loc_id:
loc_data = measurement_df[(measurement_df.Location == location)]
print(loc_data.head(1))
Date Time Location PM10 PM2.5 PM1
1593250 2020-10-19 10:29:55 535 1.96 -0.79 -0.79
Uniqueness
The data is not very unique. There are similar versions of fine dust being measured around Eindhoven, as can be seen on the samenmeten platform of the RIVD. However, there also aren’t a lot of datasets like it, with the exception of the wide scale similar measurements that samenmeten shows. This, in combination with the difference in measurement techniques makes the Uniqueness of the data moderate.
Timeliness
It took quite a while to get the data from the API, so someone who wants to collect the data from scratch the timeliness will be bad. However, now that we have all the data, the only factor is how quick the API can get a small amount of data, namely the current data.
import requests as request
start_time = time.time()
res = request.get("https://api.dustmonitoring.nl/v1/project(30)/observations?$expand=data&$filter=location%20eq%20521")
data = res.json()
print("--- %s seconds ---" % (time.time() - start_time))
--- 0.5507380962371826 seconds ---
It looks like it doesn’t take long to get the data from the API if you aren’t doing it in bulk. This is fortunate as it means that the Timeliness of the data (after it is already collected) is excellent.
Compare with report
Comparing with ILM2 report
Subdivide known location IDS into airport, city and outdoor.
airport_loc = ['I02', 'I14', 'I25']
outdoors_loc = ['I18', 'I45', 'I42', 'I43', 'I33', 'I03', 'I46', 'I47', 'I58', 'I13', 'I56', 'I10', 'I05', 'I41', 'I16', 'I44'] # 4 extra? I55 I54
city_loc = ['I51', 'I49', 'I52', 'I48', 'I04', 'I32', 'I37', 'I23', 'I07', 'I22', 'I28', 'I40', 'I08', 'I30', 'I19', 'I36', 'I39', 'I29', 'I12', 'I09', 'I11', 'I24'] # 'I57'
divided_loc_df = pd.DataFrame()
for airport in airport_loc:
loc = airboxes[airboxes['Name'] == airport]
dict_loc = {'ID': loc['ID'].values, 'Name': loc['Name'].values, 'Group': ['Airport']}
divided_loc_df = pd.concat([divided_loc_df, pd.DataFrame(dict_loc)])
for outdoor in outdoors_loc:
loc = airboxes[airboxes['Name'] == outdoor]
dict_loc = {'ID': loc['ID'].values, 'Name': loc['Name'].values, 'Group': ['Outdoors']}
divided_loc_df = pd.concat([divided_loc_df, pd.DataFrame(dict_loc)])
for city in city_loc:
loc = airboxes[airboxes['Name'] == city]
dict_loc = {'ID': loc['ID'].values, 'Name': loc['Name'].values, 'Group': ['City']}
divided_loc_df = pd.concat([divided_loc_df, pd.DataFrame(dict_loc)])
print(divided_loc_df)
ID Name Group
0 521 I02 Airport
0 536 I14 Airport
0 525 I25 Airport
0 532 I18 Outdoors
0 564 I45 Outdoors
0 561 I42 Outdoors
0 562 I43 Outdoors
0 558 I33 Outdoors
0 542 I03 Outdoors
0 565 I46 Outdoors
0 566 I47 Outdoors
0 577 I58 Outdoors
0 542 I13 Outdoors
0 575 I56 Outdoors
0 554 I10 Outdoors
0 553 I05 Outdoors
0 560 I41 Outdoors
0 556 I16 Outdoors
0 563 I44 Outdoors
0 570 I51 City
0 568 I49 City
0 571 I52 City
0 567 I48 City
0 543 I04 City
0 540 I32 City
0 528 I37 City
0 524 I23 City
0 545 I07 City
0 549 I22 City
0 526 I28 City
0 530 I40 City
0 532 I08 City
0 551 I30 City
0 547 I19 City
0 527 I36 City
0 529 I39 City
0 539 I29 City
0 535 I12 City
0 533 I09 City
0 534 I11 City
0 537 I24 City
Expected: 99% uptime.
import time
df_2021_observation = pd.DataFrame()
df_2021 = pd.DataFrame()
fmt = '%Y-%m-%d %H:%M:%S'
for location in measurement_df.Location.unique():
loc_data = measurement_df[(measurement_df.Location == location)]
d1 = datetime.strptime(loc_data.head(1).Date.values[0] + ' ' + loc_data.head(1).Time.values[0], fmt)
if d1.date().year < 2021:
d1 = datetime.strptime('2021-01-01 00:00:00', fmt)
# d2 = datetime.strptime(loc_data.tail(1).Date.values[0] + ' ' + loc_data.tail(1).Time.values[0], fmt)
d2 = datetime.strptime('2021-12-31 23:59:59', fmt)
d1_ts = time.mktime(d1.timetuple())
d2_ts = time.mktime(d2.timetuple())
mask = (loc_data['Date'] > str(d1.date())) & (loc_data['Date'] <= str(d2.date()))
loc_2021_df = loc_data.loc[mask]
df_2021 = pd.concat([df_2021, loc_2021_df])
supposed_sets = int(int(d2_ts - d1_ts) / 600)
sets_data = {'loc_id': [location], 'supposed_sets': [supposed_sets], 'actual_sets': [len(loc_2021_df)],
'percentage_missing': [100 - (
(len(loc_2021_df) / supposed_sets) * 100)]}
df_2021_observation = pd.concat([df_2021_observation, pd.DataFrame(sets_data)])
print(df_2021_observation)
loc_id supposed_sets actual_sets percentage_missing
0 521 49601 44812 9.655047
0 522 52559 47230 10.139082
0 524 52559 51913 1.229095
0 525 52559 51315 2.366864
0 526 52559 51836 1.375597
0 527 52559 52039 0.989364
0 528 49601 48689 1.838673
0 529 52559 52033 1.000780
0 530 52559 51989 1.084496
0 531 39827 29089 26.961609
0 532 52559 42347 19.429593
0 533 52559 52034 0.998877
0 534 52559 51977 1.107327
0 535 52559 52015 1.035027
0 536 52559 52039 0.989364
0 537 52559 51886 1.280466
0 539 52559 49667 5.502388
0 540 52559 50091 4.695675
0 542 52559 48943 6.879887
0 543 52559 51194 2.597081
0 544 52559 52033 1.000780
0 545 52559 50657 3.618790
0 546 52559 39450 24.941494
0 547 49731 48451 2.573847
0 549 52559 49382 6.044636
0 551 52559 52035 0.996975
0 553 52559 51170 2.642744
0 554 49601 44265 10.757848
0 556 52559 50650 3.632109
0 557 49443 44112 10.782113
0 558 52559 46832 10.896326
0 560 52559 52026 1.014098
0 561 49601 42667 13.979557
0 562 52559 51059 2.853936
0 563 49440 42496 14.045307
0 564 52559 51899 1.255732
0 565 49601 46377 6.499869
0 566 52559 41651 20.753820
0 567 49601 45314 8.642971
0 568 49601 45303 8.665148
0 569 35285 32408 8.153606
0 570 39827 34514 13.340196
0 571 39827 32084 19.441585
0 572 40378 24960 38.184160
0 573 35285 29656 15.952955
0 574 52559 21573 58.954699
0 575 52559 26243 50.069446
0 576 35285 34958 0.926739
0 577 35285 27579 21.839308
plt.hist(df_2021_observation.percentage_missing, bins=50)
plt.suptitle('Distribution of Percentage of Missing Timestamps in 2021')
plt.xlabel('Percentage missing')
plt.ylabel('Amount of locations')
plt.show()
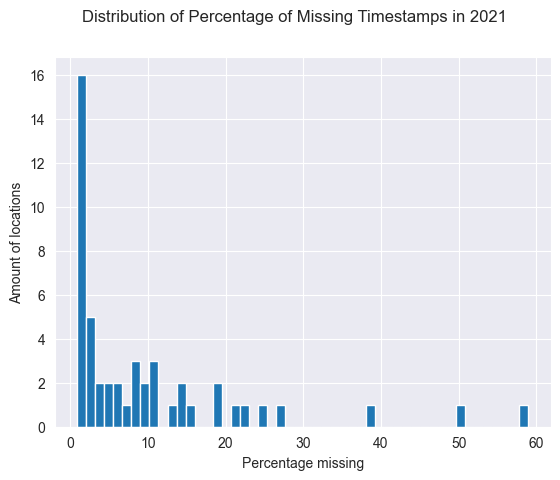
Result: about half of the locations come close to this 99% uptime.
Yearly average per amount
ilm2_df = df_2021[(df_2021.Location == 521)]
print('For ILM2:')
print('PM10 year mean: ' + str(ilm2_df['PM10'].mean()))
print('PM2.5 year mean: ' + str(ilm2_df['PM2.5'].mean()))
print('PM1 year mean: ' + str(ilm2_df['PM1'].mean()))
print('\nOverall:')
print('PM10 year mean: ' + str(df_2021['PM10'].mean()))
print('PM2.5 year mean: ' + str(df_2021['PM2.5'].mean()))
print('PM1 year mean: ' + str(df_2021['PM1'].mean()))
For ILM2:
PM10 year mean: 20.534862313665986
PM2.5 year mean: 14.855719226992772
PM1 year mean: 12.831332009283232
Overall:
PM10 year mean: 18.318628310984085
PM2.5 year mean: 12.777047338836285
PM1 year mean: 10.463851545963406
Expected ILM: PM10: 24,5 PM2.5: 18,7
Result ILM: PM10: 20,5 PM2.5: 14,8
Expected Total: PM10: 11,5 PM2.5: 13,9 PM1: 19,3
Result Total: PM10: 18,3 PM2.5: 12,8 PM1: 10.5
df_2021
| Date | Time | Location | PM10 | PM2.5 | PM1 | |
|---|---|---|---|---|---|---|
| 66 | 2021-01-22 | 00:00:00 | 521 | 6.42 | 3.42 | 2.77 |
| 67 | 2021-01-22 | 00:10:01 | 521 | 6.53 | 2.67 | 2.15 |
| 68 | 2021-01-22 | 00:20:00 | 521 | 7.15 | 2.93 | 2.03 |
| 69 | 2021-01-22 | 00:30:00 | 521 | 6.84 | 2.52 | 1.95 |
| 70 | 2021-01-22 | 00:40:00 | 521 | 6.32 | 2.60 | 2.06 |
| ... | ... | ... | ... | ... | ... | ... |
| 5667539 | 2021-12-31 | 23:10:04 | 577 | 23.53 | 16.54 | 11.50 |
| 5667540 | 2021-12-31 | 23:20:04 | 577 | 26.89 | 20.32 | 14.93 |
| 5667541 | 2021-12-31 | 23:30:04 | 577 | 24.43 | 18.48 | 12.45 |
| 5667542 | 2021-12-31 | 23:40:04 | 577 | 25.76 | 20.31 | 15.27 |
| 5667543 | 2021-12-31 | 23:50:04 | 577 | 32.12 | 24.04 | 17.81 |
2194942 rows × 6 columns
divided_loc_df = divided_loc_df.reset_index(drop=True)
print(divided_loc_df)
ID Name Group
0 521 I02 Airport
1 536 I14 Airport
2 525 I25 Airport
3 532 I18 Outdoors
4 564 I45 Outdoors
5 561 I42 Outdoors
6 562 I43 Outdoors
7 558 I33 Outdoors
8 542 I03 Outdoors
9 565 I46 Outdoors
10 566 I47 Outdoors
11 577 I58 Outdoors
12 542 I13 Outdoors
13 575 I56 Outdoors
14 554 I10 Outdoors
15 553 I05 Outdoors
16 560 I41 Outdoors
17 556 I16 Outdoors
18 563 I44 Outdoors
19 570 I51 City
20 568 I49 City
21 571 I52 City
22 567 I48 City
23 543 I04 City
24 540 I32 City
25 528 I37 City
26 524 I23 City
27 545 I07 City
28 549 I22 City
29 526 I28 City
30 530 I40 City
31 532 I08 City
32 551 I30 City
33 547 I19 City
34 527 I36 City
35 529 I39 City
36 539 I29 City
37 535 I12 City
38 533 I09 City
39 534 I11 City
40 537 I24 City
Monthly average per group.
groups_2021 = pd.DataFrame()
for location in divided_loc_df['ID']:
loc_specific = df_2021[(df_2021.Location == location)]
# if divided_loc_df[(divided_loc_df.ID == location)].Group.values == ['Airport']:
# loc_specific['Group'] = 'Airport'
print(str(location) + ' ' + divided_loc_df[(divided_loc_df.ID == location)].Group.values)
# if divided_loc_df[(divided_loc_df.ID == location)].Group.values == ['Outdoors']:
# loc_specific['Group'] = 'Outdoors'
# if divided_loc_df[(divided_loc_df.ID == location)].Group.values == ['City']:
# loc_specific['Group'] = 'City'
# groups_2021 = pd.concat([groups_2021, loc_specific])
# groups_2021_airport = pd.concat(groups_2021_airport, df_2021[(df_2021.Location == location) and (divided_loc_df[location]['Group'] == 'Airport')])
# groups_2021_outdoors = df_2021[(df_2021.Location == location.ID) and (location['Group'] == 'Outdoors')]
# groups_2021_city = df_2021[(df_2021.Location == location.ID) and (location['Group'] == 'City')]
groups_2021
# print(groups_2021_airport)
# print('For ILM2:')
# print('PM10 year mean: ' + str(ilm2_df['PM10'].mean()))
# print('PM2.5 year mean: ' + str(ilm2_df['PM2.5'].mean()))
# print('PM1 year mean: ' + str(ilm2_df['PM1'].mean()))
#
# print('\nOverall:')
# print('PM10 year mean: ' + str(df_2021['PM10'].mean()))
# print('PM2.5 year mean: ' + str(df_2021['PM2.5'].mean()))
# print('PM1 year mean: ' + str(df_2021['PM1'].mean()))
['521 Airport']
['536 Airport']
['525 Airport']
['532 Outdoors' '532 City']
['564 Outdoors']
['561 Outdoors']
['562 Outdoors']
['558 Outdoors']
['542 Outdoors' '542 Outdoors']
['565 Outdoors']
['566 Outdoors']
['577 Outdoors']
['542 Outdoors' '542 Outdoors']
['575 Outdoors']
['554 Outdoors']
['553 Outdoors']
['560 Outdoors']
['556 Outdoors']
['563 Outdoors']
['570 City']
['568 City']
['571 City']
['567 City']
['543 City']
['540 City']
['528 City']
['524 City']
['545 City']
['549 City']
['526 City']
['530 City']
['532 Outdoors' '532 City']
['551 City']
['547 City']
['527 City']
['529 City']
['539 City']
['535 City']
['533 City']
['534 City']
['537 City']
All airbox locations are unique
print(len(airboxes.ID))
print(len(airboxes.ID.unique()))
print(len(airboxes.Name.unique()))
print(len(airboxes.Description.unique()))
42
40
42
40
There are two IDs that are used twice, and have a ‘unique’ name and the lon lat combo also doesn’t match.
Now with more insights:
- 75% of relevant period needs to be measured
- background noise is measured by each hour the lowest average over the entire ILM2
- ‘Weekgang’ where you show an average week, measures its averages per hour.
Things that are odd based on previous ideas:
- Negative values should be automatically excluded
- There should be about 35 days of no data max, in 1 location, there are only 8 locations that have 1+ day of no data.
‘Claims’ that we want to verify/compare:
- The average over a week of PM2.5, PM10 and NO2 with their 25/75 percentile
- (maybe: Wind direction plot measured based on eindhoven airport weather station)
- Negative values are only mentioned on NO2, is this correct
- 8 locations have 1+ day of no data, max of 35 days.
- Fig 10, profile of the month of june in 10 min measurements
- ILM037 has an average of 20.9 ug/m3 PM10
df_521_NO2 = measurement_521_data[
(measurement_521_data.Name == 'NO2')].copy()
df_521_NO2['Timestamp'] = pd.to_datetime(df_521_NO2['Timestamp'])
df_521_NO2['Date'] = [d.date() for d in df_521_NO2['Timestamp']]
df_521_NO2['Time'] = [d.time() for d in df_521_NO2['Timestamp']]
df_521_NO2 = df_521_NO2.set_index('Timestamp')
df_521_NO2['Weekday'] = df_521_NO2.index.weekday
df_521_PM10['Weekday'] = df_521_PM10.index.weekday
df_521_PM25['Weekday'] = df_521_PM25.index.weekday
df_521_PM1['Weekday'] = df_521_PM1.index.weekday
# df = df.groupby([df['date_UTC'].dt.hour, df['w_day']]).pm2_5_ugm3.mean().reset_index().dropna()
weekday_hourly_average_PM10_521 = df_521_PM10.groupby([df_521_PM10['Weekday'], df_521_PM10.index.hour]).Value.mean()
weekday_hourly_average_PM25_521 = df_521_PM25.groupby([df_521_PM25['Weekday'], df_521_PM25.index.hour]).Value.mean()
weekday_hourly_average_PM1_521 = df_521_PM1.groupby([df_521_PM1['Weekday'], df_521_PM1.index.hour]).Value.mean()
weekday_hourly_average_NO2_521 = df_521_NO2.groupby([df_521_NO2['Weekday'], df_521_NO2.index.hour]).Value.mean()
weekday_hourly_75quantile_PM10_521 = df_521_PM10.groupby([df_521_PM10['Weekday'], df_521_PM10.index.hour]).Value.quantile(q=0.75)
weekday_hourly_25quantile_PM10_521 = df_521_PM10.groupby([df_521_PM10['Weekday'], df_521_PM10.index.hour]).Value.quantile(q=0.25)
weekday_hourly_75quantile_PM25_521 = df_521_PM25.groupby([df_521_PM10['Weekday'], df_521_PM25.index.hour]).Value.quantile(q=0.75)
weekday_hourly_25quantile_PM25_521 = df_521_PM25.groupby([df_521_PM10['Weekday'], df_521_PM25.index.hour]).Value.quantile(q=0.25)
weekday_hourly_75quantile_NO2_521 = df_521_NO2.groupby([df_521_NO2['Weekday'], df_521_NO2.index.hour]).Value.quantile(q=0.75)
weekday_hourly_25quantile_NO2_521 = df_521_NO2.groupby([df_521_NO2['Weekday'], df_521_NO2.index.hour]).Value.quantile(q=0.25)
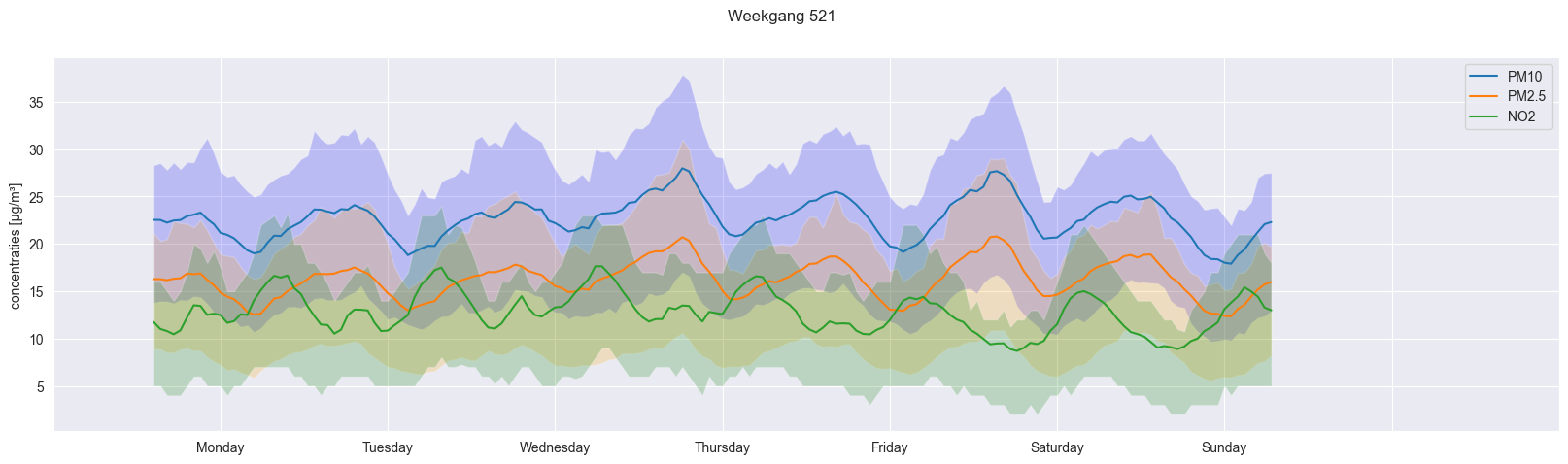
combined_dataframe = pd.DataFrame({'mean_PM10': weekday_hourly_average_PM10_521, 'mean_PM25': weekday_hourly_average_PM25_521, 'mean_NO2': weekday_hourly_average_NO2_521})
combined_dataframe_reset = combined_dataframe.reset_index(drop=True)
fig, ax = plt.subplots(figsize=(20,5))
ax.plot(combined_dataframe_reset)
ax.fill_between(combined_dataframe_reset.index, weekday_hourly_25quantile_PM10_521, weekday_hourly_75quantile_PM10_521, facecolor='blue', alpha=0.2)
ax.fill_between(combined_dataframe_reset.index, weekday_hourly_25quantile_PM25_521, weekday_hourly_75quantile_PM25_521, facecolor='orange', alpha=0.2)
ax.fill_between(combined_dataframe_reset.index, weekday_hourly_25quantile_NO2_521, weekday_hourly_75quantile_NO2_521, facecolor='green', alpha=0.2)
ax.set_xticks(ax.get_xticks() + 10,["","Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday", "", ""])
fig.suptitle("Weekgang 521")
ax.legend(['PM10', 'PM2.5', 'NO2'])
ax.set_ylabel("concentraties [µg/m\xB3]")
# fig.show()
Text(0, 0.5, 'concentraties [µg/m³]')
data = df_521_PM10.resample('60min').mean().ffill()
res = STL(data['Value'], period=24).fit()
res.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
df_2021['Datetime'] = pd.to_datetime(df_2021['Date'] + ' ' + df_2021['Time'])
df_2021 = df_2021.set_index('Datetime')
data = df_2021.resample('60min').mean().ffill()
res = STL(data['PM10'].iloc[data.index > "2021-09-01"], period=24).fit()
res.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
res = STL(data['PM10'].iloc[data.index > "2021-09-01"], period=168).fit()
res.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
Weather Data
Info
BRON: KONINKLIJK NEDERLANDS METEOROLOGISCH INSTITUUT (KNMI) Opmerking: door stationsverplaatsingen en veranderingen in waarneemmethodieken zijn deze tijdreeksen van uurwaarden mogelijk inhomogeen! Dat betekent dat deze reeks van gemeten waarden niet geschikt is voor trendanalyse. Voor studies naar klimaatverandering verwijzen we naar de gehomogeniseerde dagreeksen http://www.knmi.nl/nederland-nu/klimatologie/daggegevens of de Centraal Nederland Temperatuur http://www.knmi.nl/kennis-en-datacentrum/achtergrond/centraal-nederland-temperatuur-cnt.
SOURCE: ROYAL NETHERLANDS METEOROLOGICAL INSTITUTE (KNMI) Comment: These time series are inhomogeneous because of station relocations and changes in observation techniques. As a result, these series are not suitable for trend analysis. For climate change studies we refer to the homogenized series of daily data http://www.knmi.nl/nederland-nu/klimatologie/daggegevens or the Central Netherlands Temperature http://www.knmi.nl/kennis-en-datacentrum/achtergrond/centraal-nederland-temperatuur-cnt.
YYYYMMDD = datum (YYYY=jaar,MM=maand,DD=dag) / date (YYYY=year,MM=month,DD=day) HH = tijd (HH=uur, UT.12 UT=13 MET, 14 MEZT. Uurvak 05 loopt van 04.00 UT tot 5.00 UT / time (HH uur/hour, UT. 12 UT=13 MET, 14 MEZT. Hourly division 05 runs from 04.00 UT to 5.00 UT DD = Windrichting (in graden) gemiddeld over de laatste 10 minuten van het afgelopen uur (360=noord, 90=oost, 180=zuid, 270=west, 0=windstil, 990=veranderlijk) / Mean wind direction (in degrees) for the 10-minute period preceding the observation time stamp (360=north, 90=east, 180=south, 270=west, 0=calm, 990=variable) FH = Windsnelheid (in 0.1 m/s) gemiddeld over het afgelopen uur / Mean wind speed (in 0.1 m/s) for the hour preceding the observation time stamp FF = Windsnelheid (in 0.1 m/s) gemiddeld over de laatste 10 minuten van het afgelopen uur / Mean wind speed (in 0.1 m/s) for the 10-minute period preceding the observation time stamp “FX = Hoogste windstoot (3 seconde gemiddelde windsnelheid; in 0.1 m/s) gemeten in het afgelopen uur / Maximum wind gust (3 second mean wind speed; in 0.1 m/s) in the preceding hour” T = Temperatuur (in 0.1 graden Celsius) op 1.50 m hoogte tijdens de waarneming / Temperature (in 0.1 degrees Celsius) at 1.50 m at the time of observation T10N = Minimumtemperatuur (in 0.1 graden Celsius) op 10 cm hoogte in de afgelopen 6 uur / Minimum temperature (in 0.1 degrees Celsius) at 0.1 m inthe preceding 6-hour period TD = Dauwpuntstemperatuur (in 0.1 graden Celsius) op 1.50 m hoogte tijdens de waarneming / Dew point temperature (in 0.1 degrees Celsius) at 1.50 m at the time of observation SQ = Duur van de zonneschijn (in 0.1 uren) per uurvak, berekend uit globale straling (-1 for <0.05 uur) / Sunshine duration (in 0.1 hour) during the hourly division, calculated from global radiation (-1 for <0.05 hour) Q = Globale straling (in J/cm2) per uurvak / Global radiation (in J/cm2) during the hourly division DR = Duur van de neerslag (in 0.1 uur) per uurvak / Precipitation duration (in 0.1 hour) during the hourly division RH = Uursom van de neerslag (in 0.1 mm) (-1 voor <0.05 mm) / Hourly precipitation amount (in 0.1 mm) (-1 for <0.05 mm) P = Luchtdruk (in 0.1 hPa) herleid naar zeeniveau, tijdens de waarneming / Air pressure (in 0.1 hPa) reduced to mean sea level, at the time of observation VV = Horizontaal zicht tijdens de waarneming (0=minder dan 100m, 1=100-200m, 2=200-300m,…, 49=4900-5000m, 50=5-6km, 56=6-7km, 57=7-8km, …, 79=29-30km, 80=30-35km, 81=35-40km,…, 89=meer dan 70km) / Horizontal visibility at the time of observation (0=less than 100m, 1=100-200m, 2=200-300m,…, 49=4900-5000m, 50=5-6km, 56=6-7km, 57=7-8km, …, 79=29-30km, 80=30-35km, 81=35-40km,…, 89=more than 70km) N = Bewolking (bedekkingsgraad van de bovenlucht in achtsten), tijdens de waarneming (9=bovenlucht onzichtbaar) / Cloud cover (in octants), at the time of observation (9=sky invisible) U = Relatieve vochtigheid (in procenten) op 1.50 m hoogte tijdens de waarneming / Relative atmospheric humidity (in percents) at 1.50 m at the time of observation WW = Weercode (00-99), visueel(WW) of automatisch(WaWa) waargenomen, voor het actuele weer of het weer in het afgelopen uur. Zie https://cdn.knmi.nl/knmi/pdf/bibliotheek/scholierenpdf/weercodes_Nederland.pdf / Present weather code (00-99), description for the hourly division. IX = Weercode indicator voor de wijze van waarnemen op een bemand of automatisch station (1=bemand gebruikmakend van code uit visuele waarnemingen, 2,3=bemand en weggelaten (geen belangrijk weersverschijnsel, geen gegevens), 4=automatisch en opgenomen (gebruikmakend van code uit visuele waarnemingen), 5,6=automatisch en weggelaten (geen belangrijk weersverschijnsel, geen gegevens), 7=automatisch gebruikmakend van code uit automatische waarnemingen) / Indicator present weather code (1=manned and recorded (using code from visual observations), 2,3=manned and omitted (no significant weather phenomenon to report, not available), 4=automatically recorded (using code from visual observations), 5,6=automatically omitted (no significant weather phenomenon to report, not available), 7=automatically set (using code from automated observations) M = Mist 0=niet voorgekomen, 1=wel voorgekomen in het voorgaande uur en/of tijdens de waarneming / Fog 0=no occurrence, 1=occurred during the preceding hour and/or at the time of observation R = Regen 0=niet voorgekomen, 1=wel voorgekomen in het voorgaande uur en/of tijdens de waarneming / Rainfall 0=no occurrence, 1=occurred duringthe preceding hour and/or at the time of observation S = Sneeuw 0=niet voorgekomen, 1=wel voorgekomen in het voorgaande uur en/of tijdens de waarneming / Snow 0=no occurrence, 1=occurred during the preceding hour and/or at the time of observation O = Onweer 0=niet voorgekomen, 1=wel voorgekomen in het voorgaande uur en/of tijdens de waarneming / Thunder 0=no occurrence, 1=occurred during the preceding hour and/or at the time of observation Y = IJsvorming 0=niet voorgekomen, 1=wel voorgekomen in het voorgaande uur en/of tijdens de waarneming / Ice formation 0=no occurrence, 1=occurred during the preceding hour and/or at the time of observation
eindhoven_weather_hourly = pd.read_csv('csv_files/uurgeg_370_2021-2030.csv', skiprows=31, sep='\,', engine='python')
for item in eindhoven_weather_hourly['"# STN'].index:
eindhoven_weather_hourly['"# STN'][item] = int(eindhoven_weather_hourly['"# STN'][item][1:])
eindhoven_weather_hourly[' Y"'][item] = int(eindhoven_weather_hourly[' Y"'][item][:-1])
eindhoven_weather_hourly = eindhoven_weather_hourly.rename(columns={'"# STN': '# STN', ' Y"': 'Y'})
columns = []
for col in eindhoven_weather_hourly.columns:
columns.append(col.strip())
eindhoven_weather_hourly.columns = columns
eindhoven_weather_hourly_relevant = eindhoven_weather_hourly[['YYYYMMDD','HH', 'DD', 'FH', 'RH', 'T']]
eindhoven_weather_hourly_relevant.columns = ['Date', 'Hour', 'WindDir', 'WindSpeed_avg', 'PrecipationHour', 'Temp']
# eindhoven_weather_hourly_relevant.to_csv(df_file_path + 'hourdata_eindhoven_2021_16may2023.csv', index=False)
eindhoven_weather_hourly_relevant.tail(1)
| Date | Hour | WindDir | WindSpeed_avg | PrecipationHour | Temp | |
|---|---|---|---|---|---|---|
| 20783 | 20230516 | 24 | 290 | 20 | 0 | 67 |
eindhoven_weather_hourly = pd.read_csv('csv_files/uurgeg_370_2021-2030.csv', skiprows=31, skipinitialspace=True)
eindhoven_weather_hourly.tail()
| # STN,YYYYMMDD, HH, DD, FH, FF, FX, T, T10N, TD, SQ, Q, DR, RH, P, VV, N, U, WW, IX, M, R, S, O, Y | |
|---|---|
| 20779 | 370,20230516, 20, 330, 40, 40, 70, 1... |
| 20780 | 370,20230516, 21, 320, 20, 20, 40, ... |
| 20781 | 370,20230516, 22, 300, 20, 20, 30, ... |
| 20782 | 370,20230516, 23, 300, 20, 20, 30, ... |
| 20783 | 370,20230516, 24, 290, 20, 20, 30, ... |
eindhoven_weather_hourly_full = pd.read_csv('csv_files/hourdata_eindhoven_2021_16may2023.csv')
eindhoven_weather_hourly_full
| Date | Hour | WindDir | WindSpeed_avg | PrecipationHour | Temp | |
|---|---|---|---|---|---|---|
| 0 | 20210101 | 1 | 290 | 10 | 0 | 20 |
| 1 | 20210101 | 2 | 250 | 10 | 0 | 18 |
| 2 | 20210101 | 3 | 220 | 10 | 0 | 11 |
| 3 | 20210101 | 4 | 270 | 20 | 0 | 10 |
| 4 | 20210101 | 5 | 250 | 20 | 0 | 3 |
| ... | ... | ... | ... | ... | ... | ... |
| 20779 | 20230516 | 20 | 330 | 40 | 0 | 105 |
| 20780 | 20230516 | 21 | 320 | 20 | 0 | 90 |
| 20781 | 20230516 | 22 | 300 | 20 | 0 | 78 |
| 20782 | 20230516 | 23 | 300 | 20 | 0 | 71 |
| 20783 | 20230516 | 24 | 290 | 20 | 0 | 67 |
20784 rows × 6 columns
weather_measurements_by_datetime = eindhoven_weather_hourly_relevant.copy()
datetimes = []
for index in weather_measurements_by_datetime.index:
string_to_convert = str(weather_measurements_by_datetime['Date'][index]) + " " + str(weather_measurements_by_datetime['Hour'][index] - 1)
datetimes.append(datetime.strptime(string_to_convert, "%Y%m%d %H"))
weather_measurements_by_datetime['DateTime'] = datetimes
print(weather_measurements_by_datetime.tail())
Date Hour WindDir WindSpeed_avg PrecipationHour Temp \
20779 20230516 20 330 40 0 105
20780 20230516 21 320 20 0 90
20781 20230516 22 300 20 0 78
20782 20230516 23 300 20 0 71
20783 20230516 24 290 20 0 67
DateTime
20779 2023-05-16 19:00:00
20780 2023-05-16 20:00:00
20781 2023-05-16 21:00:00
20782 2023-05-16 22:00:00
20783 2023-05-16 23:00:00
measurement_df = pd.read_csv(df_file_path + 'full_measurement.csv')
measurement_df.tail()
| Date | Time | Location | PM10 | PM2.5 | PM1 | |
|---|---|---|---|---|---|---|
| 5739474 | 2023-05-19 | 10:10:03 | 577 | 11.65 | 6.02 | 4.69 |
| 5739475 | 2023-05-19 | 10:20:03 | 577 | 13.06 | 6.85 | 4.92 |
| 5739476 | 2023-05-19 | 10:30:03 | 577 | 11.15 | 7.33 | 4.76 |
| 5739477 | 2023-05-19 | 10:40:03 | 577 | 11.88 | 6.35 | 4.47 |
| 5739478 | 2023-05-19 | 10:50:03 | 577 | 14.06 | 7.43 | 5.20 |
measurement_df['DateTime'] = pd.to_datetime(measurement_df['Date'] + ' ' + measurement_df['Time'])
measurement_df = measurement_df.set_index('DateTime', drop=True)
grouper = measurement_df.groupby([pd.Grouper(freq='1H'), 'Location'])\
# .mean().ffill().asfreq('1H')
# hourly_measurement_full.sample(20)
result = grouper.mean().ffill()
hourly_measurement_full = result.reset_index()
weather_measurements_by_datetime
| Date | Hour | WindDir | WindSpeed_avg | PrecipationHour | Temp | DateTime | |
|---|---|---|---|---|---|---|---|
| 0 | 20210101 | 1 | 290 | 10 | 0 | 20 | 2021-01-01 00:00:00 |
| 1 | 20210101 | 2 | 250 | 10 | 0 | 18 | 2021-01-01 01:00:00 |
| 2 | 20210101 | 3 | 220 | 10 | 0 | 11 | 2021-01-01 02:00:00 |
| 3 | 20210101 | 4 | 270 | 20 | 0 | 10 | 2021-01-01 03:00:00 |
| 4 | 20210101 | 5 | 250 | 20 | 0 | 3 | 2021-01-01 04:00:00 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 20779 | 20230516 | 20 | 330 | 40 | 0 | 105 | 2023-05-16 19:00:00 |
| 20780 | 20230516 | 21 | 320 | 20 | 0 | 90 | 2023-05-16 20:00:00 |
| 20781 | 20230516 | 22 | 300 | 20 | 0 | 78 | 2023-05-16 21:00:00 |
| 20782 | 20230516 | 23 | 300 | 20 | 0 | 71 | 2023-05-16 22:00:00 |
| 20783 | 20230516 | 24 | 290 | 20 | 0 | 67 | 2023-05-16 23:00:00 |
20784 rows × 7 columns
combine_weather_measurements = hourly_measurement_full.copy()
combine_weather_measurements['WindDir'] = np.nan
combine_weather_measurements['WindSpeed_avg'] = np.nan
combine_weather_measurements['PrecipationHour'] = np.nan
combine_weather_measurements['Temp'] = np.nan
print(len(weather_measurements_by_datetime.index))
combine_weather_measurements_full = pd.DataFrame()
for index in weather_measurements_by_datetime.index:
relevant_data = combine_weather_measurements[combine_weather_measurements['DateTime'] == weather_measurements_by_datetime['DateTime'][index]].copy()
relevant_data['WindDir'] = weather_measurements_by_datetime['WindDir'][index]
relevant_data['WindSpeed_avg'] = weather_measurements_by_datetime['WindSpeed_avg'][index]
relevant_data['PrecipationHour'] = weather_measurements_by_datetime['PrecipationHour'][index]
relevant_data['Temp'] = weather_measurements_by_datetime['Temp'][index]
combine_weather_measurements_full = pd.concat([combine_weather_measurements_full, relevant_data])
remaining = combine_weather_measurements[combine_weather_measurements['DateTime'] >= '2023-05-17 00:00:00']
import requests as request
for date in remaining['DateTime'].map(lambda t: t.date()).unique():
relevant_data = combine_weather_measurements[combine_weather_measurements['DateTime'].dt.date == date].copy()
relevant_data['WindDir'] = weather_measurements_by_datetime['WindDir'][index]
url = 'http://api.weatherapi.com/v1/history.json?key=2bd0e4d77ef74cf5bec123551231805&q=Eindhoven&dt=' + str(date)
res = request.get(url)
data = res.json()
for hour in range(len(data['forecast']['forecastday'][0]['hour'])):
relevant_hour = relevant_data[relevant_data['DateTime'].dt.hour == hour].copy()
relevant_hour['WindDir'] = data['forecast']['forecastday'][0]['hour'][hour]['wind_degree']
relevant_hour['WindSpeed_avg'] = data['forecast']['forecastday'][0]['hour'][hour]['wind_kph'] / 3.6
relevant_hour['PrecipationHour'] = data['forecast']['forecastday'][0]['hour'][hour]['precip_mm']
relevant_hour['Temp'] = data['forecast']['forecastday'][0]['hour'][hour]['temp_c']
combine_weather_measurements_full = pd.concat([combine_weather_measurements_full, relevant_hour])
# combine_weather_measurements_full.to_csv(df_file_path + 'hour_combined_data_eindhoven_2021_19may2023.csv')
current_data = combine_weather_measurements_full[combine_weather_measurements_full['Location'] == 521]
if pd.to_datetime(current_data.tail(1)['DateTime'].values[0]).date() < datetime.now().date():
print(pd.to_datetime(current_data.tail(1)['DateTime'].values[0]).date())
print(datetime.now().date())
elif pd.Timestamp(current_data.tail(1)['DateTime'].values[0]).hour < datetime.now().hour:
print(pd.Timestamp(current_data.tail(1)['DateTime'].values[0]).hour)
print(datetime.now().hour)
2023-05-16
2023-06-05
last_datetime = current_data.tail(1).DateTime.values[0]
pd.to_datetime(last_datetime).strftime("%Y-%m-%dT%H:%M:%SZ")
'2023-05-16T23:00:00Z'
fig, ax = plt.subplots(figsize=(20,5))
ax.plot(combined_dataframe_reset)
ax.fill_between(combined_dataframe_reset.index, weekday_hourly_25quantile_PM10_521, weekday_hourly_75quantile_PM10_521, facecolor='blue', alpha=0.2)
ax.fill_between(combined_dataframe_reset.index, weekday_hourly_25quantile_PM25_521, weekday_hourly_75quantile_PM25_521, facecolor='orange', alpha=0.2)
ax.fill_between(combined_dataframe_reset.index, weekday_hourly_25quantile_NO2_521, weekday_hourly_75quantile_NO2_521, facecolor='green', alpha=0.2)
ax.set_xticks(ax.get_xticks() + 10,["","Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday", "", ""])
fig.suptitle("Weekgang 521")
ax.legend(['PM10', 'PM2.5', 'NO2'])
ax.set_ylabel("concentraties [µg/m\xB3]")
# fig.show()
Text(0, 0.5, 'concentraties [µg/m³]')
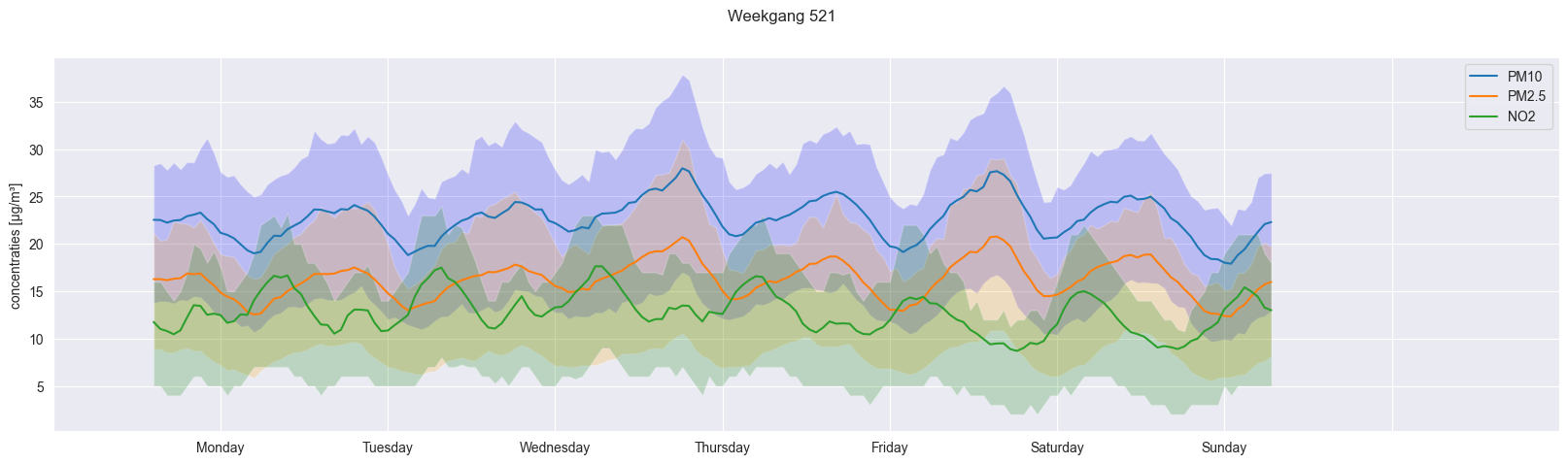
data = df_521_PM10.resample('60min').mean().ffill()
res = STL(data['Value'], period=24).fit()
res.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
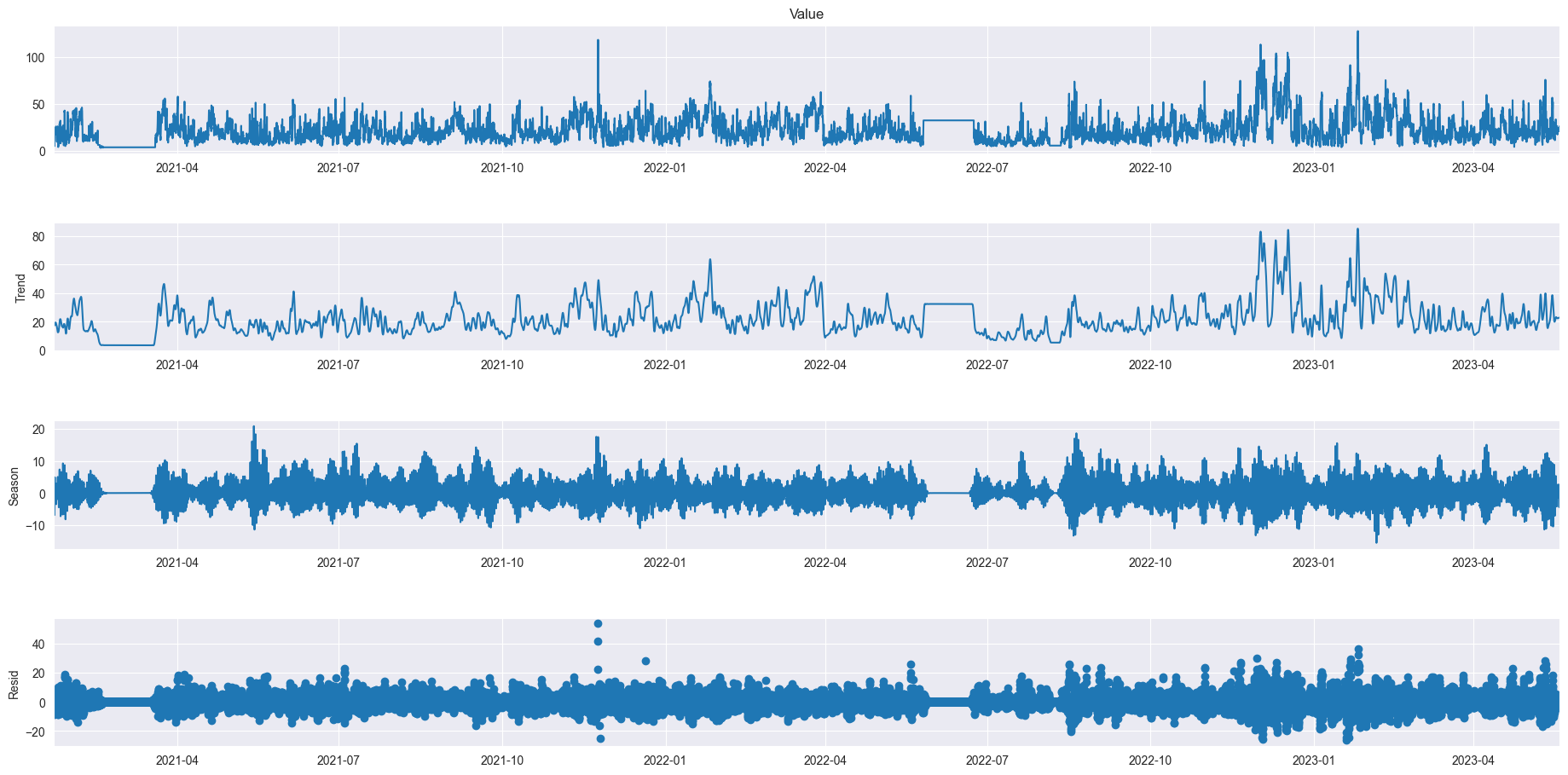
df_2021['Datetime'] = pd.to_datetime(df_2021['Date'] + ' ' + df_2021['Time'])
df_2021 = df_2021.set_index('Datetime')
data = df_2021.resample('60min').mean().ffill()
res = STL(data['PM10'].iloc[data.index > "2021-09-01"], period=24).fit()
res.plot()
plt.gcf().set_size_inches(20,10)
plt.show()
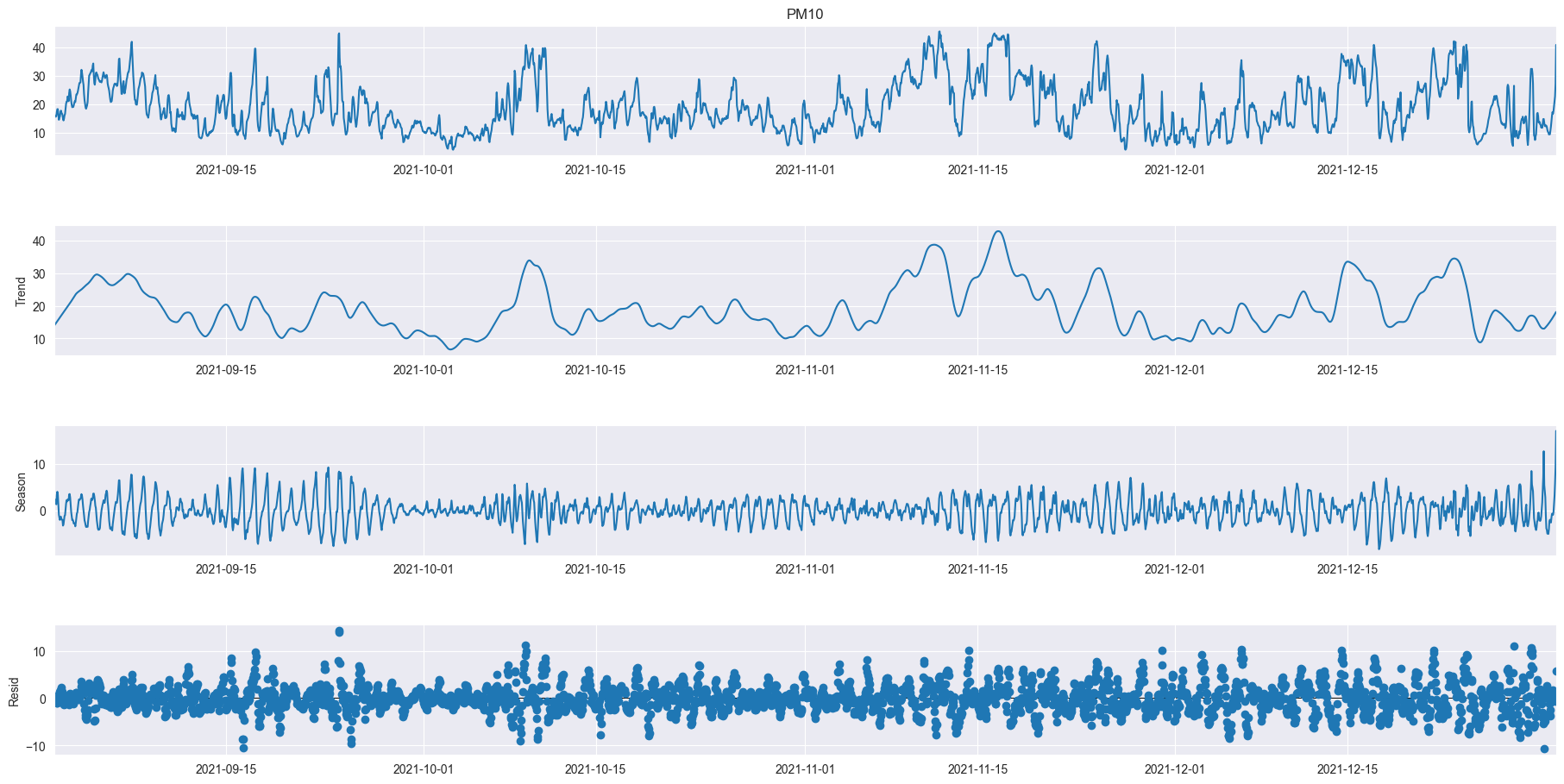
res = STL(data['PM10'].iloc[data.index > "2021-09-01"], period=168).fit()
res.plot()
plt.gcf().set_size_inches(20,10)
plt.show()